 | ||
In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by differences in the non-recombining portions of DNA from the Y chromosome (called Y-DNA). It represents human genetic diversity based on single-nucleotide polymorphisms (SNPs) on the Y chromosome.
Contents
- Naming convention
- Major Y DNA haplogroups
- Haplogroups A B
- Haplogroup CT P143
- Haplogroup C M130
- Haplogroup F M89
- Haplogroup D M174
- Haplogroup E M96
- Haplogroup G M201
- Haplogroup H M69
- Haplogroup I M170
- Haplogroup J M304
- Haplogroup K M9
- Haplogroups L T K1
- Haplogroup K2 K M526
- Haplogroup N K2a1
- Haplogroup O K2a2
- Haplogroups K2b1 M S
- Haplogroup P K2b2
- Haplogroup Q M242
- Haplogroup R M207
- References
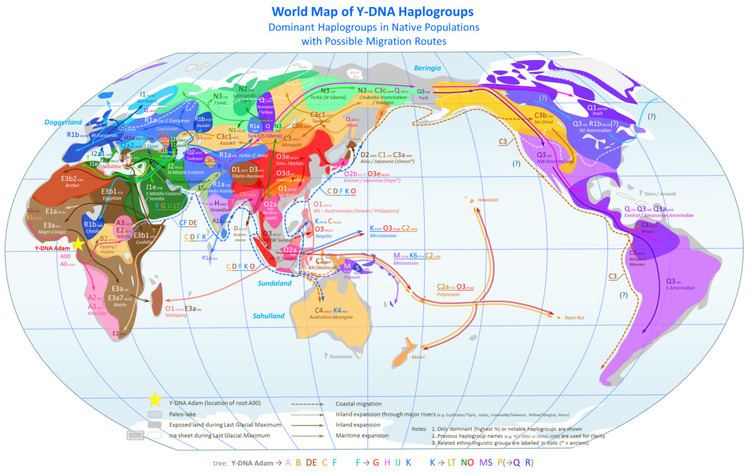
Y-DNA haplogroups represent major branches of the Y-chromosome Phylogenetic tree. Y-chromosomal Adam is the name given by researchers to the patrilineal most recent common ancestor of all living humans at the root of this tree. Estimates of the date when Y-chromosomal Adam lived have varied significantly in different studies. Archaeological and genetic data suggest that the source populations of Paleolithic humans survived the glacial maxima (including the LGM) and human Y-DNA haplogroups emerged in sparsely wooded refugia, and dispersed through areas of high primary productivity while avoiding dense forest cover.
Naming convention
Y-DNA haplogroups are defined by the presence of a series of Y-DNA SNP markers. Subclades are defined by a terminal SNP, the SNP furthest down in the Y-chromosome phylogenetic tree. The Y Chromosome Consortium (YCC) developed a system of naming major Y-DNA haplogroups with the capital letters A through T, with further subclades named using numbers and lower case letters (YCC longhand nomenclature). YCC shorthand nomenclature names Y-DNA haplogroups and their subclades with the first letter of the major Y-DNA haplogroup followed by a dash and the name of the defining terminal SNP.
Y-DNA haplogroup nomenclature is changing over time to accommodate the increasing number of SNPs being discovered and tested, and the resulting expansion of the Y-chromosome phylogenetic tree. This change in nomenclature has resulted in inconsistent nomenclature being used in different sources. This inconsistency, and increasingly cumbersome longhand nomenclature, has prompted a move towards using the simpler shorthand nomenclature. In September 2012, Family Tree DNA provided the following explanation of its changing Y-DNA haplogroup nomenclature to individual customers on their Y-DNA results pages (note that the haplogroup mentioned below relates to a specific individual):
Major Y-DNA haplogroups
Major Y-chromosome haplogroups include:
Haplogroups A & B
Using fast evolving SNPs, Haplogroup A is the macrohaplogroup from which all modern paternal haplogroups descend. It is sparsely distributed in Africa. BT is a Subclade of Haplogroup A; more precisely of A1b (A2-T in Cruciani et al. 2011), as follows:
Haplogroup CT (P143)
The defining mutations separating CT (all haplogroups excepting A and B) are M168 and M294. These mutations predate the "Out of Africa" migration. The defining mutations of DE probably occurred in Northeastern Africa some 65,000 years ago. The P143 mutation that defines Haplogroup CF may have occurred at that time, bringing modern humans to the southern coast of Asia.
Subclades:
Haplogroup C (M130)
Haplogroup F (M89)
The groups descending from haplogroup F are found in some 90% of the world's population, but almost exclusively outside of sub-Saharan Africa.
F xG,H,I,J,K is rare in modern populations and peaks in South Asia, especially Sri Lanka. It also appears to have long been present in South East Asia. has been reported at rates of 4-5% in Sulawesi and Lembata. One study, which did not comprehensively screen for other subclades of F-M89 (including some subclades of GHIJK), found that Indonesian men with the SNP P14/PF2704 (which is equivalent to M89), comprise 1.8% of men in West Timor, 1.5% of Flores 5.4% of Lembata 2.3% of Sulawesi and 0.2% in Sumatra. F* (F xF1,F2,F3) has been reported among 10% of males in Sri Lanka and South India, 5% in Pakistan, as well as lower levels among the Tamang people (Nepal), and in Iran. F1 (P91), F2 (M427) and F3 (M481; previously F5) are all highly rare and virtually exclusive to regions/ethnic minorities in Sri Lanka, India, Nepal, South China, Thailand, Burma, and Vietnam.In such cases, however, the possibility of misidentification is considered to be relatively high and some may belong to misidentified subclades of Haplogroup GHIJK.
Haplogroup D (M174)
Haplogroup E (M96)
Haplogroup G (M201)
Haplogroup G (M201) originated in the Middle East or further east – possibly even the Wardak region of Afghanistan some 30,000 years BP. It spread to Europe with the Neolithic Revolution.
It is found in many ethnic groups in Eurasia; most common in the Caucasus, Iran, Anatolia and the Levant. Found in almost all European countries, but most common in Gagauzia, southeastern Romania, Greece, Italy, Spain, Portugal, Tyrol, and Bohemia with highest concentrations on some Mediterranean islands; uncommon in Northern Europe.
G-M201 is also found in small numbers in northwestern China and India, Pakistan, Sri Lanka, Malaysia, and North Africa.
Haplogroup H (M69)
Haplogroup H (M69) probably emerged in South Asia, about 30,000 to 40,000 years BP, and remains prevalent there, in the forms of H1 (M69) and H3 (Z5857).
However, H2 (P96) has been present in Europe since the Neolithic and H1a1 (M82) spread westward in the Medieval era with the migration of the Romani .
Haplogroup I (M170)
Haplogroup I (M170, M258) is found mainly in Europe and the Caucasus.
Haplogroup J (M304)
Haplogroup J (M304, S6, S34, S35) is found mainly in the Middle East and South-East Europe.
Haplogroup K (M9)
Haplogroup K (M9) is spread all over Eurasia, Oceania and among Native Americans'
Haplogroups L & T (K1)
Haplogroup L (M20) is found in South Asia, Central Asia, South-West Asia, and the Mediterranean.
Haplogroup T (M184, M70, M193, M272) is found in the Middle East, Africa (mainly Afro-Asiatic-speaking peoples), the Mediterranean, and South Asia. Found in a significant minority of Sciaccensi, Somalis, Stilfser, Ethiopians, Fulbe, Egyptians, Oman, Sephardi Jews, and Eivissencs, is; also found at low frequency throughout the Mediterranean and parts of India
Haplogroup K2 (K-M526)
The only living males reported to carry the basal paragroup K2* are indigenous Australians. Major studies published in 2014 and 2015 suggest that up to 27% of Aboriginal Australian males carry K2*, while others carry a subclade of K2.
Haplogroup N (K2a1)
Haplogroup N (M231) also known as K2a1 is found through northern Eurasia, especially among the Uralic peoples.
Haplogroup N possibly originated in eastern Asia and spread both west into Siberia and north, being the most common group found in some Uralic speaking peoples. Haplogroup O is found at its highest frequency in East Asia and Southeast Asia, with lower frequencies in the South Pacific, Central Asia, and South Asia.
Haplogroup O (K2a2)
Haplogroup O (M175) is found in East Asia, Southeast Asia, and the South Pacific.
Haplogroups K2b1, M & S
NO examples of the basal paragroup K2b1* have been identified. Males carrying subclades of K2b1 are found primarily among Papuan peoples, Micronesian peoples, indigenous Australians, and Polynesians.
Its subclades include two major haplogroups
Haplogroup P (K2b2)
Haplogroup P (P295) has two primary branches: P1 (P-M45) and the extremely rare P2 (P-B253).
P*, P1* and P2 are found together only on the island of Luzon, in The Philippines. In particular, P* and P1* are found at significant rates among members of the Aeta (or Agta) people of Luzon. While, P1* is now more common among living individuals in Eastern Siberia and Central Asia, it is also found at low levels in mainland South East Asia and South Asia. Considered together, these distributions tend to suggest that P* emerged from K2b in South East Asia.
P1 is also the parent node of two primary clades:
Haplogroup Q (MEH2, M242, P36) found in Siberia and the Americas Haplogroup R (M207, M306): found in Europe, West Asia, Central Asia, and South Asia
Haplogroup Q M242
Q is defined by the SNP M242. It is believed to have arisen in Central Asia approximately 17,000 to 22,000 years ago. The subclades of Haplogroup Q with their defining mutation(s), according to the 2008 ISOGG tree are provided below. ss4 bp, rs41352448, is not represented in the ISOGG 2008 tree because it is a value for an STR. This low frequency value has been found as a novel Q lineage (Q5) in Indian populations
The 2008 ISOGG tree
Haplogroup R (M207)
Haplogroup R is defined by the SNP M207. The bulk of Haplogroup R is represented in descendant subclade R1, which likely originated on the Eurasian Steppes. R1 has two descendant subclades: R1a and R1b.
R1a is associated with the proto-Indo-Iranian and Balto-Slavic peoples, and is now found primarily in Central Asia, South Asia, and Eastern Europe.
Haplogroup R1b is the dominant haplogroup of Western Europe and also found sparsely distributed among various peoples of Asia and Africa. Its subclade R1b1a2 (M269) is the haplogroup that is most commonly found among modern Western European populations, and has been associated with the Italo-Celtic and Germanic peoples.
