Defining mutations M231 | Ancestor NO | |
 | ||
Possible time of origin 20,000 to 25,000 years BP | ||
Haplogroup N (M231) is a Y-chromosome DNA haplogroup typical of Northern Eurasia, which is defined by the presence of the marker M231.
Contents
- Origins
- Distribution
- N M231
- N1 CTS11499L735M2291
- N1c1 M46
- N1c1a M178
- N1c2a M128
- N1c2b P43
- Phylogenetic history
- Original research publications
- Associated mutations SNPs and UEPs
- Tree
- References
However, the basal paragroup N* has only been found in populations indigenous to China and Cambodia. Subclades of N-M231 have been found at low levels in Southeast Asia, the Pacific Islands, Southwest Asia and the Balkans. These factors tend to suggest that it originated in East Asia or Southeast Asia.
Origins
Haplogroup N-M231 is a descendant haplogroup of Haplogroup NO. It is considered relatively young, having populated the north of Eurasia after the last Ice Age. Males carrying the marker apparently moved northwards as the climate warmed in the Holocene.
It is suggested that N-M231 arose in Southeast Asia 19.4±4.8 ky years ago, and then migrated in a counter-clockwise path from modern day regions of Mongolia and northern China to as far as northeastern Europe (Rootsi 2006).
The absence of haplogroup N-M231 in the Americas indicates that its spread across Asia happened after the submergence of Beringia (Chiaroni 2009).
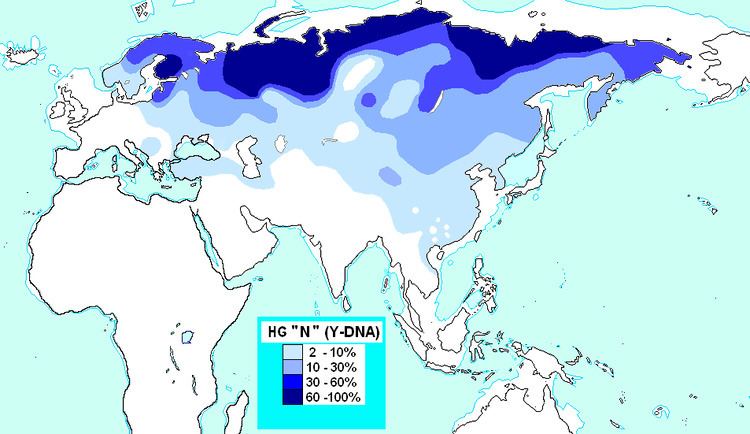
Distribution
Haplogroup N has a wide geographic distribution throughout northern Eurasia, and it also has been observed occasionally in other areas, including Southeast Asia, the Pacific, Southwest Asia and Southern Europe.
Its highest frequency occurs among the Finnic and Baltic peoples of northern Europe, the Ob-Ugric and Northern Samoyedic peoples of western Siberia, and the Siberian Turkic-speaking Yakuts (McDonald 2005).
N* (M231)
Y-chromosomes that display the M231 mutation that defines Haplogroup N-M231, but do not display the CTS11499, L735, M2291 mutations that define Haplogroup N1 are said to belong to paragroup N-M231*. (A "Haplogroup N2" has also been mooted, defined by F3373, M2283, Page56, and/or S323.)
N-M231* has been found at low levels in China and Cambodia. Out of a sample of 165 Han males from China, two individuals (1.2%) were found to belong to N*.(Karafet 2010). One originated from Guangdong and one from Shaanxi.
N1 (CTS11499/L735/M2291)
In 2014, LLY22g was retired as a defining SNP for Haplogroup N1; it was replaced by CTS11499/L735/M2291. According to ISOGG, LLY22g is problematic because it is a "palindromic marker and can easily be misinterpreted". Consequently, the position of many previously examples of "N-LLY22g", within N-M231 has become unclear.
N1* has been reported to reach a frequency of up to 30% (13/43) among the Yi people of Butuo County, Sichuan in Southwest China (Hammer 2005, Karafet 2001, and Wen2004b). Paragroup N-LLY22g* also has been found in samples of Han Chinese, but with widely varying frequency:
Other populations in which representatives of N1 * have been found include:
N1c1 (M46)
The mutations that define the subclade N-M46 are M46/Tat and P105. This is the most frequent subclade of N. It arose probably in the region of present-day China, and subsequently experienced serial bottlenecks in Siberia and secondary expansions in eastern Europe (Rootsi 2006). Haplogroup N-M46 is approximately 14,000 years old.
In Siberia, haplogroup N-M46 reaches a maximum frequency of approximately 90% among the Yakuts, a Turkic people who live mainly in the Sakha (Yakutia) Republic. However, it is practically non-existent among many of the Yakuts' neighboring ethnic groups, such as Tungusic speakers. It also has been detected in 5.9% (3/51) of a sample of Hmong Daw from Laos (Cai 2011), 2.4% (2/85) of a sample from Seoul, South Korea (Katoh 2004), and in 1.4% (1/70) of a sample from Tokushima, Japan (Hammer 2005).
The haplogroup N-M46 has a low diversity among Yakuts suggestive of a population bottleneck or founder effect ( & Pakendorf 2002). This was confirmed by a study of ancient DNA which traced the origins of the male Yakut lineages to a small group of horse-riders from the Cis-Baikal area (Crubézy 2010).
N1c1a (M178)
The subclade N-M178 is defined by the presence of markers M178 and P298. N-M178* has higher average frequency in Northern Europe than in Siberia, reaching frequencies of approximately 60% among Finns and approximately 40% among Latvians, Lithuanians & 35% among Estonians (Derenko 2007 and Lappalainen 2008).
Miroslava Derenko and her colleagues noted that there are two subclusters within this haplogroup, both present in Siberia and Northern Europe, with different histories. The one that they labelled N3a1 first expanded in south Siberia (approximately 10,000 years ago on their calculated by the Zhivotovsky method) and spread into Northern Europe where its age they calculated as around 8,000 years ago. Meanwhile, the younger subcluster, which they labelled N3a2, originated in south Siberia (probably in the Baikal region) approximately 4,000 years ago (Derenko 2007).
N1c2a-M128
This subclade is defined by the presence of the marker M128. N-M128 was first identified in a sample from Japan (1/23 = 4.3%) and in a sample from Central Asia and Siberia (1/184 = 0.5%) in a preliminary survey of worldwide Y-DNA variation. Subsequently, it has been found with low frequency in some samples of the Manchu people, Sibe people, Evenks, Koreans, northern Han Chinese, Bouyei people, and some Turkic peoples of Central Asia.
A number of Han Chinese, an Ooled Mongol, a Qiang, and a Tibetan were found to belong to a sister branch (or branches) of N-M128 under paragroup N-F1154*.
N1c2b (P43)
Haplogroup N-P43 is defined by the presence of the marker P43. It is a significantly younger subclade, perhaps only 6,000 to 8,000 years old, with a probable origin in Siberia (Derenko 2007). It is found frequently among Northern Samoyedic peoples; also found at low to moderate frequency among some other Uralic peoples, Turkic peoples, Mongolic peoples, Tungusic peoples, and Siberian Yupik people.
Haplogroup N-P43 forms two distinctive subclusters of STR haplotypes, Asian and European, the latter mostly distributed among Uralic-speaking peoples and related populations (Rootsi 2006).
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
Original research publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Associated mutations (SNPs and UEPs)
B1/B3 The b2/b3 deletion in the AZFc region of the Y-chromosome. This deletion appears to have occurred independently on at least four different occasions. Therefore, this deletion should not be taken as a unique event polymorphism defining this branch of the Y-chromosome tree (ISOGG 2012).
Tree
In the following tree the nomenclature of 3 sources is separated by slashes: ISOGG (2016)/Ilumae et al. (2016)/Kang Hu et al. (2015).
