Possible time of origin 53,000 years BP [1] Ancestor CF | Possible place of origin Asia | |
 | ||
Descendants C1 F3393/Z1426 (previously CxC3)C2 (previously C3*) M217. Defining mutations M130/RPS4Y711, P184, P255, P260 | ||
Haplogroup C is a major Y-chromosome haplogroup, defined by UEPs M130/RPS4Y, P184, P255, and P260, which are all SNP mutations. One of two primary branches of Haplogroup CF alongside Haplogroup F. Haplogroup C is found in ancient populations on every continent except Africa and is the predominant Y-DNA haplogroup among males belonging to many peoples indigenous to East Asia, Central Asia, Siberia, North America and Oceania. It is also found at moderate frequencies among certain indigenous populations of South Asia and Southeast Asia.
Contents
In addition to the basal paragroup C*, this haplogroup now has two major branches: C1 (F3393/Z1426; previously CxC3, i.e. old C1, old C2, old C4, old C5 and old C6) and C2 (M217; the former C3).
Origins
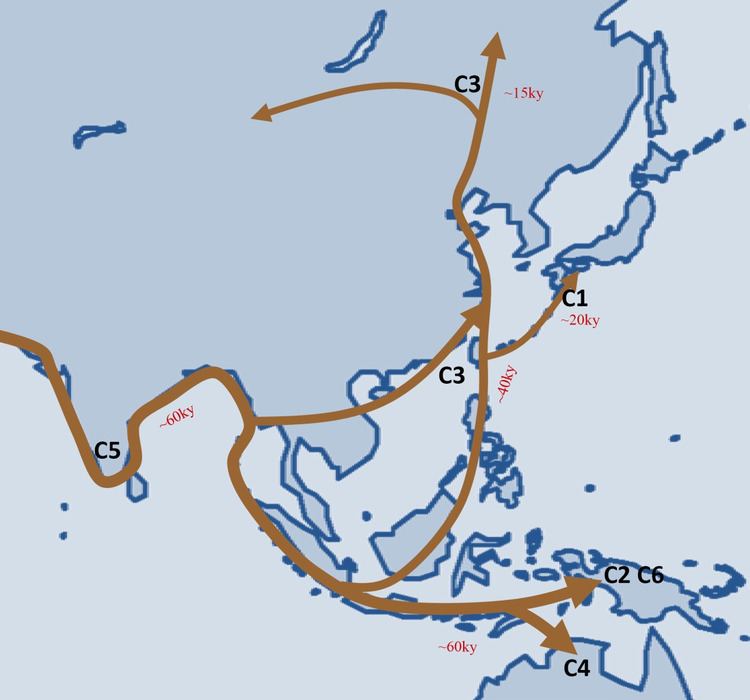
Haplogroup C-M130 seems to have come into existence shortly after SNP mutation M168 occurred for the first time, bringing the modern Haplogroup CT into existence, from which Haplogroup CF, and in turn Haplogroup C, derived. This was probably at least 60,000 years before present.
Although Haplogroup C-M130 attains its highest frequencies among the indigenous populations of Mongolia, the Russian Far East, Polynesia, Australia, and at moderate frequency in Korea and Manchu people, it displays its highest diversity among modern populations of India. It is therefore hypothesized that Haplogroup C-M130 either originated or underwent its longest period of evolution within India or the greater South Asian coastal region. The highest diversity is observed in Southeast Asia, and its northward expansion in East Asia started approximately 40,000 years ago.
Males carrying C-M130 are believed to have migrated to the Americas some 6,000-8,000 years before present, and was carried by Na-Dené-speaking peoples into the northwest Pacific coast of North America.
Asia is also the area in which Haplogroup D-M174 is concentrated. However, D-M174 is more closely related to haplogroup E than to C-M130 and the geographical distributions of Haplogroups C-M130 and D-M174 are entirely and utterly different, with various subtypes of Haplogroup C-M130 being found at high frequency amongst indigenous Australians, Polynesians, Vietnamese, Kazakhs, Mongolians, Manchurians, Koreans, and indigenous inhabitants of the Russian Far East; and at moderate frequencies elsewhere throughout Asia and Oceania, including India, Sri Lanka and Southeast Asia. Whereas Haplogroup D is found at high frequencies only amongst Tibetans, Japanese peoples, and Andaman Islanders, and has been found neither in India nor among the aboriginal inhabitants of the Americas or Oceania.
Structure
C* (M130/Page51/RPS4Y711, M216)
(The above phylogenetic structure of haplogroup C-M130 subclades is based on the ISOGG 2015 tree, YCC 2008 tree and subsequent published research.)
Distribution
The distribution of Haplogroup C-M130 is generally limited to populations of northern Asia, eastern Asia, Oceania, and the Americas. Due to the tremendous age of Haplogroup C, numerous secondary mutations have had time to accumulate, and many regionally important subbranches of Haplogroup C-M130 have been identified.
Up to 46% of Aboriginal Australian males carried either basal C* (C-M130*), C1b2b* (C-M347*) or C1b2b1 (C-M210), before contact with and significant immigration by Europeans, according to a 2015 study by Nagle et al. That is, 20.0% of the Y-chromosomes of 657 modern individuals, before 56% of those samples were excluded as "non-indigenous". C-M130* was apparently carried by up to 2.7% of Aboriginal males before colonisation; 43% carried C-M347, which has not been found outside Australia. (The other haplogroups regarded as preceding contact with Europeans among Aboriginal males subclades of K, namely basal K*, K2b1* and subclades of K2b1.)
Low levels of C-M130* are carried by males:
C* (M130) was also identified in prehistoric remains, dating from 34,000 years BP, found in Russia and known as "Kostenki 14".
Haplogroup C-M217 – the most numerous and widely dispersed C lineage – which is believed to have originated in South East/Central Asia, spread from there into Northern Asia and the Americas. C-M217 stretches longitudinally from Central Europe and Turkey, to the Wayuu people of Colombia and Venezuela, and latitudinally from the Athabaskan peoples of Alaska to Vietnam to the Malay Archipelago. Found at low concentrations in Eastern Europe, where it may be a legacy of the invasions/migrations of the Huns, Turks and/or Mongols during the Middle Ages. Found at especially high frequencies in Buryats, Daurs, Hazaras, Itelmens, Kalmyks, Koryaks, Manchus, Mongolians, Oroqens, and Sibes, with a moderate distribution among other Tungusic peoples, Koreans, Ainus, Nivkhs, Altaians, Tuvinians, Uzbeks, Han Chinese, Tujia, Hani, and Hui. The highest frequencies of Haplogroup C-M217 are found among the populations of Mongolia and Far East Russia, where it is the modal haplogroup. Haplogroup C-M217 is the only variety of Haplogroup C-M130 to be found among Native Americans, among whom it reaches its highest frequency in Na-Dené populations.
Other subclades are specific to certain populations, within a restricted geographical range; even where these other branches are found, they tend to appear as a very low-frequency, minor component of the palette of Y-chromosome diversity within those territories:
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
Research publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Notable members
One particular haplotype within Haplogroup C-M217 has received a great deal of attention for the possibility that it may represent direct patrilineal descent from Genghis Khan.
