Descendants D, E | Ancestor CT | |
 | ||
Possible time of origin 65,000 (59,100–68,300) BP 50,000-55,000 split between D and E, 70,000-75,000 split between CF and DE Defining mutations M1/YAP, M145 = P205, M203, P144, P153, P165, P167, P183 | ||
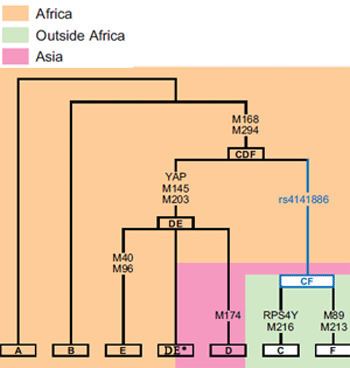
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183.
Contents
DE is unique because it is distributed in several geographically distinct clusters. Immediate subclade, haplogroup D, is normally found only in eastern Asia, and the other immediate subclade, haplogroup E, is common in Africa, Europe and the Middle East.
The most well-known unique event polymorphism (UEP) that defines DE is the Y-chromosome Alu Polymorphism (YAP) YAP. The mutation was caused when a strand of DNA, known as Alu, inserted a copy of itself into the Y chromosome. Hence all Y chromosomes belonging to DE, D, E and their subclades are YAP-positive (YAP+). All Y chromosomes that belong to other haplogroups and subclades are YAP-negative (YAP-).
Haplogroup DE is an estimated 65,000 years old.
Distribution
The subclades of DE continue to confound investigators trying to reconstruct the migration of humans because, while they are common in Africa and East Asia, they are also largely absent between these two regions. As the paragroup DE* is extremely rare, the majority of DE male lines fall into subclades of either D-M174 or E-M96. While D-M174 likely originated in Asia – the only place where it is now found, E-M96 is more likely to have originated in East Africa. However, a West Asian origin for E-M96 is considered possible by some scholars. Some researchers have suggested that the exceptional rarity of DE lineages in India – a region considered important in the dispersal of modern humans – may be meaningful, because D-M174 and E-M96 have remained common in neighbouring areas. It has been inferred that a deleterious mutation or mutations may have been responsible for the extinction of DE lineages in India.
DE* has been found in three separate studies, at very low frequencies, among males from two widely separated locations: West Africa and East Asia. A 2003 study by Weale et al., of the DNA of over 8,000 males worldwide, found that five out of 1,247 Nigerian males belonged to DE*. The DE* found possessed by these five Nigerians, according to the study's authors, was "the least derived of all YAP chromosomes according to currently known binary markers" – to such an extent that it suggested that DE had originated in West Africa and expanded from there. However, Weale et al., cautioned that such inferences may well be incorrect. In addition, the seemingly "paraphyletic" (basal) status of the Nigerian examples of DE-YAP may be "illusory" because the "branching order, and hence the origin, of YAP-derived haplogroups remains uncertain". It was "easy to misinterpret apparently paraphyletic groups", and subsequent research might show that the Nigerian examples of DE were as divergent from DE*, D* and E*. "[T]he only genealogically meaningful definition of the age of a clade is the time to its most recent common ancestor, but only if DE* is [truly] paraphyletic does it ... become automatically older than D or E..." The relationship between DE*, according to Weale et al., "can be viewed as a missing-data problem..." Nevertheless, in 2007, another West African example of DE* was reported – carried by a speaker of the Nalu language who was among 17 Y-DNA samples taken in Guinea Bissau. The sequence of this individual differed by one mutation from those of the Nigerian individuals, indicating common ancestry, although the relationship between the two lineages has not been determined. In 2008, DE* was identified in two individuals from Tibet (two out of 594). It remains possible that the above examples of DE* are divergent forms of E and/or D, or the result of a back mutation.
Discovery
The YAP insertion was discovered by scientists led by Michael Hammer of the University of Arizona. Between 1997 and 1998 Hammer published three articles relating to the origins of haplogroup DE. These articles state that YAP insertion occurred in Asia. As recently as 2007, some studies such as Chandrasekar et al. 2007, cite the publications by Hammer when arguing for an Asian origin of the YAP insertion.
The scenarios outlined by Hammer include an out of Africa migration over 100,000 years ago, the YAP+ insertion on an Asian Y-chromosome 55,000 years ago and a back migration of YAP+ from Asia to Africa 31,000 years ago by its subclade haplogroup E. This analysis was based on the fact that older African lineages, such as haplogroups A and B, were YAP negative whereas the younger lineage, haplogroup E was YAP positive. Haplogroup D, which is YAP positive, was clearly an Asian lineage, being found only in East Asia with high frequencies in Andaman Islands, Japan and Tibet. Because the mutations that define haplogroup E were observed to be in the ancestral state in haplogroup D, and haplogroup D at 55kya, was considerably older than haplogroup E at 31kya, Hammer concluded that haplogroup E was a subclade of haplogroup D.
Contemporary studies
In 2000 a number of scientists had started to reassess the hypothesis of an Asian origin of the YAP insertion. Underhill et al. 2000 identified the D-M174 mutation that defines haplogroup D. The M174 allele is found in the ancestral state in all African lineages including haplogroup E. The discovery of M174 mutation meant that haplogroup E could not be a subclade of haplogroup D. These findings effectively neutralized the argument of an Asian origin of the YAP+ based on the character state of the M40 and M96 mutations that define haplogroup E. According to Underhill et al. 2000, the M174 data alone would support an African origin of the YAP insertion.
Further arguments were made supporting and African origin of the YAP in Underhill et al. 2001. The arguments for an African origin include.
- Africa has the highest frequency of YAP(>80%). Whereas the YAP+ in Asia has a fairly restricted geographic distribution, mainly at low to moderate frequencies (average 9.6%) in East Asia.
- It was claimed that there was no archaeological evidence of a back-migration to Africa, and at the time of writing that there was no unequivocal Y DNA, mitochondrial DNA or autosomal DNA evidence of a back migration to Africa.
- Although Haplogroup C seems to have originated in Asia at a similar time to Haplogroup DE's origin, Haplogroup C shows no sign of back migration to Africa.
The African origin of the YAP+ is also supported by studies concerning haplogroup E. In Altheide and Hammer 1997, the authors argue that haplogroup E arose in Asia on an ancestral YAP+ allele before migrating back to Africa. However some studies, such as Semino et al., indicate that the highest frequency and diversity of haplogroup E is in Africa, and Africa is the most likely place of origin of the haplogroup.
The models supporting an African origin or an Asian origin of the YAP+ insertion both required the extinction of the ancestral YAP chromosome to explain the current distribution of the YAP+ polymorphism. Paragroup DE* possesses neither the mutations that define haplogroup D or haplogroup E. If paragroup DE* was found in one location but not the other, it would boost one theory of the other. Haplogroup DE* has recently been found in Nigeria, Guinea-Bissau and also in Tibet. The phylogenetic relationship of three DE* sequences has yet to be determined, but it is known that the Guinea Bissau sequences differ from the Nigerian sequences by at least one mutation. Weale et al. state that the discovery of DE* among Nigerians pushes back the date for the most recent common ancestor (MRCA) of African YAP chromosomes. This, in his view, has the effect of reducing the time window through which a possible back migration from Asia to Africa could occur.
Chandrasekhar et al. 2007, have argued for the Asian origin of the YAP+. They state,
The presence of the YAP insertion in Northeast Indian tribes and Andaman Islanders with haplogroup D suggests that some of the M168 chromosomes gave rise to the YAP insertion and M174 mutation in South Asia
They also argue that YAP+ migrated back to Africa with other Eurasian haplogroups. These include Haplogroup R1b1* (18-23kya), which has been observed with especially high frequency among the members of some peoples in northern Cameroon, and Haplogroup T (39-45 kya), which has been observed in low frequencies in Africa. Haplogroup E at 50kya is considerably older than these haplogroups and has been observed at frequencies of 80-92% in Africa.
In a press release concerning a study by Karafet et al. (2008), Michael Hammer, revised the dates for the origin Haplogroup DE from 55,000 years ago to 65,000 years ago. For haplogroup E, Hammer revised the dates from 31,000 years ago to 50,000 years ago. Hammer is also quoted as saying “The age of haplogroup DE is about 65,000 years, just a bit younger than the other major lineage to leave Africa, which is assumed to be about 70,000 years old,” in which he implies that haplogroup DE left Africa along with Haplogroup CF.
Peter Underhill states that there will always be uncertainty regarding the precise origins of DNA sequence variants such as YAP because of a lack of knowledge concerning prehistoric demographics and population movements. However Underhill contends that with all the available information, the African origin of the YAP+ polymorphism is more parsimonious and more plausible than the Asian origin hypothesis. Other authors who have published or co-published works in support of an African origin of the YAP+ include Luigi Luca Cavalli-Sforza, Toomas Kivisild, Spencer Wells, Linda Stone and Paul F. Lurquin.
Tree
This phylogenetic tree of para-haplogroup DE is based on the YCC 2008 tree and subsequent published research.
