Ancestor R1 | ||
 | ||
Descendants One immediate descendant – R1b1 (L278, M415, P25).Three secondary descendants – R1b1a (R-M269); R1b1b (R-P297); R1b1c (R-V88) Defining mutations 1. M343 defines R1b in the broadest sense2. In some cases, major downstream mutations such as M269 – which now defines R1b1a2 – are used to identify R1b, especially in regional or out-of-date studies. | ||
Haplogroup R1b (R-M343), known initially as Hg1 and Eu18, is a human Y-chromosome haplogroup.
Contents
- Origin and dispersal
- External phylogeny
- Internal structure
- R1b R M343
- R1b1 R L278
- R1b1a1 R L388
- R1b1a2 R V88
- R1b1b R M335
- In popular culture
- References
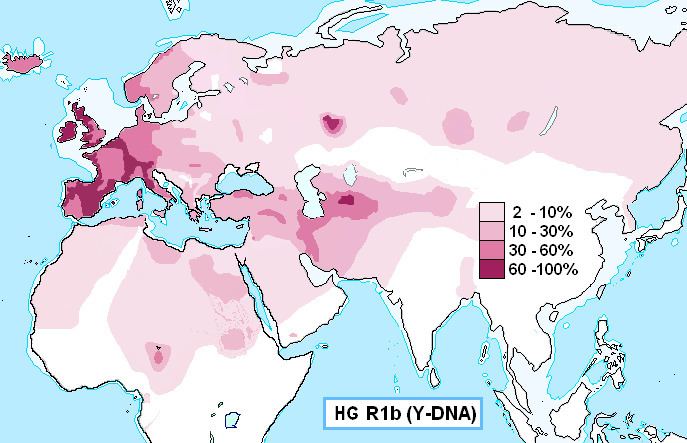
It is the most frequently occurring paternal lineage in Western Europe, as well as some parts of Russia (e.g. the Bashkir minority) and Central Africa (e.g. Chad and Cameroon). R1b also reaches high frequencies in the Americas and Australasia, due largely to immigration from Western Europe. There is an ongoing debate regarding the origins of R1b subclades found at significant levels among some indigenous peoples of the Americas, such as speakers of Algic languages in central Canada. It is also present at lower frequencies throughout Eastern Europe, Western Asia, as well as parts of North Africa and Central Asia.
R1b has one primary branch, R1b1 (L278), which in turn has two primary branches: R1b1a (L75) and R1b1b (PH155). R1b1a is found mostly in Western Europe, although the Ful?e and Chadic-speaking peoples of Africa are dominated by R1b1a2 (PF6279/V88). Western Europe is dominated by the downstream subclades of R1b1a – especially R1b1a1a2 (R-M269), previously known as R1b1a2. R1b1b (PH155) is seen mostly in Western and Central Asia.
Origin and dispersal
R1b is a subclade within the "macro-haplogroup" Haplogroup K (K-M9), which is one of the predominant groupings of all the rest of human male lines outside of Africa. K* is believed to have originated in Asia (as is the case with an even earlier ancestral haplogroup, F (F-M89). Karafet T. et al. (2014) "rapid diversification process of K-M526 likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q."
Three autosomal genetic studies in 2015 gave support to the Kurgan theory of Gimbutas regarding the Indo-European Urheimat. According to those studies, haplogroups R1b and R1a, now the most common in Europe (R1a is also common in South Asia) would have expanded from the Russian steppes, along with the Indo European languages; they also detected an autosomal component present in modern Europeans which was not present in Neolithic Europeans, which would have been introduced with paternal lineages R1b and R1a, as well as Indo European Languages.
The point of origin of R1b is thought to lie in Eurasia, most likely in Western Asia. T. Karafet et al. (2008) estimated the age of R1, the parent of R1b, as 18,500 years before present.
Early research into the origins of R1b focused on Europe. In 2000, Ornella Semino and colleagues argued that R1b had been in Europe before the end of the Ice Age, and had spread north from an Iberian refuge after the Last Glacial Maximum. Age estimates of R1b in Europe have steadily decreased in more recent studies, at least concerning the majority of R1b, with more recent studies suggesting a Neolithic age or younger. On the other hand, Morelli et al. have recently (in 2010) attempted to defend a Palaeolithic origin for R1b1b2. Irrespective of microsatellite coalescence calculations, Chikhi et al. pointed out that the timing of molecular divergences does not coincide with population splits; the TMRCA of haplogroup R1b (whether in the Palaeolithic or Neolithic) dates to its point of origin somewhere in Eurasia, and not its arrival in western Europe. Summing up, Michael R. Maglio argues that the closest branch of R1b is from Iberia and its small subclades found in West Asia, the Near East and Africa are examples of back migration, and not of its origin.
However, as Barbara Arredi and colleagues were the first to point out, the distribution of R1b microsatellite variance in Europe forms a cline from east to west, which is more consistent with an entry into Europe from Western Asia with the spread of farming. A 2009 paper by Chiaroni et al. added to this perspective by using R1b as an example of a wave haplogroup distribution, in this case from east to west. The proposal of a southeastern origin of R1b were supported by three detailed studies based on large datasets published in 2010. These detected that the earliest subclades of R1b are found in western Asia and the most recent in western Europe.
While age estimates in these articles are all more recent than the Last Glacial Maximum, all mention the Neolithic, when farming was introduced to Europe from the Middle East as a possible candidate period. Myres et al. (August 2010), and Cruciani et al. (August 2010) both remained undecided on the exact dating of the migration or migrations responsible for this distribution, not ruling out migrations as early as the Mesolithic or as late as Hallstatt but more probably Late Neolithic. They noted that direct evidence from ancient DNA may be needed to resolve these gene flows. Lee et al. (May 2012) analysed the ancient DNA of human remains from the Late Neolithic Beaker culture site of Kromsdorf, Germany identifying two males as belonging to the Y haplogroup R1b. Analysis of ancient Y DNA from the remains of populations derived from early Neolithic Central and North European Linear Pottery culture settlements have not yet found males belonging to haplogroup R1b.
The suggestion has also been made that the spread of R1b in Western Europe may coincide with the spread of the Centum branch of the Indo-European languages during the early Bronze Age.
One of the highest level of R1b is found among the Basques, who speak a non- Indo-European language isolate. One hypothesis about the case of the Basques is that a male-dominated Indo-European-speaking people invaded and conquered the Basque region, and then, having brought no or few women with them, then married local women, possibly from a matrilineal society The women then raised the children that resulted to speak their language and cultural practices, rather than those of their fathers. This hypothesis is supported by the fact that while other high-R1b regions in Western Europe (such as the British Isles and southern Germany) also show disproportionately high incidences of MtDNA haplogroups that correspond to a Pontic Steppes origin (specifically MtDNA Haplogroups I, U2, U3, U4, and W), while the Basque region does not. In fact, the Basque region displays virtually no MtDNA for which Pontic Steppes origin could be claimed.
External phylogeny
R1b is a part of the broader Haplogroup K-M9 and its linear descendants K2, K2b and P, which is also known as K2b2. Tatiana et al. (2014) "rapid diversification process of K-M526 likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q."
Internal structure
Names such as R1b, R1b1 and so on are phylogenetic (i.e. "family tree") names which make clear their place within the branching of haplogroups, or the phylogenetic tree. An alternative way of naming the same haplogroups and subclades refers to their defining SNP mutations: for example, R-M343 is equivalent to R1b. Phylogenetic names change with new discoveries and SNP-based names are consequently reclassified within the phylogenetic tree. In some cases, an SNP is found to be unreliable as a defining mutation and an SNP-based name is removed completely. For example, before 2005, R1b was synonymous with R-P25, which was later reclassified as R1b1; in 2016, R-P25 was removed completely as a defining SNP, due to a significant rate of back-mutation.
This is the basic outline of R1b according to the ISOGG Tree as it stood on January 30, 2017.
R1b* (R-M343*)
R1b* – that is, males with M343, but no subsequent distinguishing SNP mutations – is extremely rare. The only population yet recorded with a definite significant proportion of R1b* are the Kurds of southeastern Kazakhstan with 13%. However, more recently, a large study of Y-chromosome variation in Iran, revealed R1b* as high as 4.3% among Iranian sub-populations.
While studies in 2005–08 suggested that R1b* may occur at high levels in Jordan, Egypt and Sudan, subsequent research indicates that the samples concerned most likely belong to the subclade R1b1a2 (R-V88), which is now concentrated among African populations, following back migration from Asia. It remains a possibility that some or even most of these cases may be R1b* (R-M343*), R1a* (R-M420*), an otherwise undocumented branch of R1, and/or back-mutations of a marker, from a positive to a negative ancestral state, constituting, in other words, undocumented subclades of R1b. Thus demonstrating the importance of testing for SNPs critical in identifying subclades.
A compilation of previous studies regarding the distribution of R1b can be found in Cruciani et al. (2010). It is summarised in the table following. (It should be noted that Cruciani did not include some studies suggesting even higher frequencies of R1b1a1a2 [R-M269] in some parts of Western Europe.)
R1b1 (R-L278)
R1b1*, like R1b* is rare. However, the skeletons of two males from both a Mesolithic pre-Yamna Samara culture burial dated to around 5650–5555 BC north of the Caspian Sea and an early Neolithic Cardial culture burial dated to around 5178–5066 BCE at the Els Trocs site, Aragon, in the Pyrenees, Spain were found to contain R1b1*.
Some examples described in older articles, for example two found in Turkey, are now thought to be mostly in the more recently discovered sub-clade R1b1a2 (R-V88). Most or all examples of R1b therefore fall into subclades R1b1a2 (R-V88) or R1b1a (R-P297). Cruciani et al. in the large 2010 study found 3 cases amongst 1173 Italians, 1 out of 328 West Asians and 1 out of 156 East Asians. Varzari found 3 cases in the Ukraine, in a study of 322 people from the Dniester-Carpathian Mountains region, who were P25 positive, but M269 negative. Cases from older studies are mainly from Africa, the Middle East or Mediterranean, and are discussed below as probable cases of R1b1a2 (R-V88).
R1b1a1 (R-L388)
R-L388, also known as R1b1a1 (L388/PF6468, L389/PF6531) appears to be rare or extinct in its basal form. Its subclades are also relatively rare and found in various parts of South West Asia, the Mediterranean basin and continental Europe.
R1b1a1a (R-P297)
The SNP marker P297 was recognised in 2008 as ancestral to the significant subclades M73 and M269, combining them into one cluster. This had been given the phylogenetic name R1b1a1a (and, previously, R1b1a).
A majority of Eurasian R1b falls within this subclade, representing a very large modern population. Although P297 itself has not yet been much tested for, the same population has been relatively well studied in terms of other markers. Therefore, the branching within this clade can be explained in relatively high detail below. The skeleton of a male from a Chalcolithic Yamna burial in the Middle-Volga-Samara area, dated to around 3305–2925 BC, was found to possibly contain R1b1a* being P297 positive but L51 negative.
R1b1a1a1 (R-M73)
R-M73 is reportedly the dominant haplogroup among the Kumandin of the Altai Republic in Russia.
While early research into R-M73 claimed that it was significantly represented among the Hazara of Afghanistan and the Bashkirs of the Ural Mountains, this has apparently been overturned. For example, supporting material from a 2010 study by Behar et al. suggested that Sengupta et al. (2006) had misidentified Hazara individuals, who instead belonged to R2 or Q. Likewise, most Bashkir males have been found to belong to R-152 (R1b1a1a2a1a2b) and some, mostly from south-eastern Bashkortostan, belonged to Haplogroup Q-M25 (Q1a1b) rather than R1b.
R1b1a1a2 (R-M269)
R-M269 (previously R1b1a2, amongst other names) is defined by the presence of SNP marker M269. R1b1a2* or M269 (xL23) is found at highest frequency in the central Balkans notably Kosovo with 7.9%, Macedonia 5.1% and Serbia 4.4%. Kosovo is notable in having a high percentage of descendant L23* or L23(xM412) at 11.4% unlike most other areas with significant percentages of M269* and L23* except for Poland with 2.4% and 9.5% and the Bashkirs of southeast Bashkortostan with 2.4% and 32.2% respectively. Notably this Bashkir population also has a high percentage of M269 sister branch M73 at 23.4%. Five individuals out of 110 tested in the Ararat Valley, Armenia belonged to R1b1a2* and 36 to L23*, with none belonging to known subclades of L23.
European R1b is dominated by R-M269. It has been found at generally low frequencies throughout central Eurasia, but with relatively high frequency among the Bashkirs of the Perm region (84.0%) and Baymaksky District (81.0%). This marker is present in China and India at frequencies of less than one percent. The table below lists in more detail the frequencies of M269 in regions in Asia, Europe, and Africa.
Trofimova et al. (2015) found a surprising high frequency of R1b-L23 (Z2105/2103) among the peoples of the Idel-Ural. 21 out of 58 (36.2%) of Burzyansky District Bashkirs, 11 out of 52 (21.2%) of Udmurts, 4 out of 50 (8%) of Komi, 4 out of 59 (6.8%) of Mordvins, 2 out of 53 (3.8%) of Besermyan and 1 out of 43 (2.3%) of Chuvash were R1b-L23 (Z2105/2103), the type of R1b found in the recently analyzed Yamna remains of the Samara Oblast and Orenburg Oblast.
The frequency is about 92% in Wales, 82% in Ireland, 70% in Scotland, 68% in Spain, 60% in France (76% in Normandy), about 60% in Portugal, 53% in Italy, 45% in Eastern England, 50% in Germany, 50% in the Netherlands, 42% in Iceland, and 43% in Denmark. It is as high as 95% in parts of Ireland. It is also found in some areas of North Africa, where its frequency peaks at 10% in some parts of Algeria. M269 has likewise been observed among 8% of the Herero in Namibia.
The R-M269 subclade has been found in ancient Guanche (Bimbapes) fossils excavated in Punta Azul, El Hierro, Canary Islands, which are dated to the 10th century (~44%).
From 2003 to 2005, what is now R1b1a2 was designated R1b3. From 2005 to 2008, it was R1b1c. From 2008 to 2011, it was R1b1b2.
As discussed above, in articles published around 2000 it was proposed that this clade had been in Europe before the last ice age, but by 2010 more recent periods such as the European Neolithic have become the focus of proposals. A range of newer estimates for R1b1a2, or at least its dominant parts in Europe, are from 4,000 to a maximum of about 10,000 years ago, and looking in more detail is seen as suggesting a migration from Western Asia via southeastern Europe. Western European R1b is dominated by R-P310.
In this period between 2000 and 2010 that it became clear that especially Western European R1b is dominated by specific sub-clades of R-M269 (with some small amounts of other types found in areas such as Sardinia). Within Europe, R-M269 is dominated by R-M412, also known as R-L51, which according to Myres et al. (2010) is "virtually absent in the Near East, the Caucasus and West Asia." This Western European population is further divided between R-P312/S116 and R-U106/S21, which appear to spread from the western and eastern Rhine river basin respectively. Myres et al. note further that concerning its closest relatives, in R-L23*, it is "instructive" that these are often more than 10% of the population in the Caucasus, Turkey, and some southeast European and circum-Uralic populations. In Western Europe it is present but in generally much lower levels apart from "an instance of 27% in Switzerland's Upper Rhone Valley." In addition, the sub-clade distribution map, Figure 1h titled "L11(xU106,S116)", in Myres et al. shows that R-P310/L11* (or as yet undefined subclades of R-P310/L11) occurs only in frequencies greater than 10% in Central England with surrounding areas of England and Wales having lower frequencies. This R-P310/L11* is almost non-existent in the rest of Eurasia and North Africa with the exception of coastal lands fringing the western and southern Baltic (reaching 10% in Eastern Denmark and 6% in northern Poland) and in Eastern Switzerland and surrounds.
In 2009, DNA extracted from the femur bones of 6 skeletons in an early-medieval burial place in Ergolding (Bavaria, Germany) dated to around 670 AD yielded the following results: 4 were found to be haplogroup R1b with the closest matches in modern populations of Germany, Ireland and the USA while 2 were in Haplogroup G2a.
Population studies which test for M269 have become more common in recent years, while in earlier studies men in this haplogroup are only visible in the data by extrapolation of what is likely. The following gives a summary of most of the studies which specifically tested for M269, showing its distribution (as a percentage of total population) in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.
R-L23* (R1b1a1a2a*) is now most commonly found in Anatolia, the Caucasus and the Mediterranean .
R-L51* (R1b1a1a2a1*) is now concentrated in a geographical cluster centred on southern France and northern Italy.
R-L151 (L151/PF6542, CTS7650/FGC44/PF6544/S1164, L11, L52/PF6541, P310/PF6546/S129, P311/PF6545/S128) also known as R1b1a1a2a1, and its subclades, include most males with R1b in Western Europe.
This subclade is defined by the presence of the SNP U106, also known as S21 and M405. It appears to represent over 25% of R1b in Europe. In terms of percentage of total population, its epicenter is Friesland, where it makes up 44% of the population. In terms of total population numbers, its epicenter is Central Europe, where it comprises 60% of R1 combined.
While this sub-clade of R1b is frequently discussed amongst genetic genealogists, the following table represents the peer-reviewed findings published so far in the 2007 articles of Myres et al. and Sims et al.
Along with R-U106, R-P312 is one of the most common types of R1b1a2 (R-M269) in Europe. Also known as S116, it has been the subject of significant study concerning its sub-clades, and some of the ones recognized by the ISOGG tree as of December 27, 2015 are summarized in the following table. Myres et al. described it distributing from the west of the Rhine basin.
Amongst these, scientific publications have given interpretation and comment on several:-
R-P312-b (R-U152) is defined by the presence of the marker U152, also called S28. Its discovery was announced in 2005 by EthnoAncestry and subsequently identified independently by Sims et al. (2007). Myres et al. report this clade "is most frequent (20–44%) in Switzerland, Italy, France and Western Poland, with additional instances exceeding 15% in some regions of England and Germany." Similarly Cruciani et al. (2010) reported frequency peaks in Northern Italy and France. Out of a sample of 135 men in Tyrol, Austria, 9 tested positive for U152/S28. Far removed from this apparent core area, Myres et al. also mention a sub-population in north Bashkortostan where 71% of 70 men tested were in R-U152. They propose this to be the result of an isolated founder effect. King et al. (2014) reported four living relatives of King Richard III of England in the male line tested positive for U-152. However, DNA analysis of Richard III's skeleton showed he had a haplotype G-P287. The researchers concluded there must have been a non-paternal event in the intervening generations.
R-P312-c (R-L21) is defined by the presence of the marker L21, also referred to as M529 and S145. Myres et al. report it is most common in England and Ireland (25–50% of the whole male population). Known sub-clades include the following:-
R1b1a1b (R-V1636)
R-V1636 (R1b1a1b) has no known subclades, is rare and has been found mostly in the broader Mediterranean region.
R1b1a2 (R-V88)
R1b1a2 (PF6279/V88; previously R1b1c) is defined by the presence of SNP marker V88, the discovery of which was announced in 2010 by Cruciani et al. Apart from individuals in southern Europe and Western Asia, the majority of R-V88 was found in the Sahel among populations speaking Afroasiatic languages of the Chadic branch:
As can be seen in the above data table, R1b1c is found in northern Cameroon in west central Africa at a very high frequency, where it is considered to be caused by a pre-Islamic movement of people from Eurasia.
R1b1a2a (R-M18)
R1b1a2a is a sub-clade of R-V88, which is defined by the presence of SNP marker M18. It has been found only at low frequencies in samples from Sardinia and Lebanon.
R1b1b (R-M335)
R1b1b is defined by the presence of SNP marker M335. This haplogroup was created by the 2008 reorganisation of nomenclature and should not be confused with R1b1b2, which was previously called R1b1c. Its position in relation to the much more populous sub-clade R1b1a is uncertain. The M335 marker was first published in 2004, when one example was discovered in Turkey, which was classified at that time as R1b4.
ISOGG 2017 cites M335 as "R1b1b1", alongside "R1b1b2" (PH200), both descended from R1b1b (PH155).
In popular culture
Bryan Sykes, in his book Blood of the Isles, gives the populations associated with R1b the name of Oisín for a clan patriarch, much as he did for mitochondrial haplogroups in The Seven Daughters of Eve.
Stephen Oppenheimer also deals with this haplogroup in his book Origins of the British, giving the R1b clan patriarch the Basque name "Ruisko" in honour of what he thinks is the Iberian origin of R1b.
Artem Lukichev has created a (non-scientific) animation based on a Bashkir epic about the Ural, which outlined the history of the clusters of haplogroup R1: R1a and R1b.
The DNA tests that assisted in the identification of Czar Nicholas II of Russia found that he belonged to R1b.
