Possible time of origin 39,800-45,500 years BP | ||
 | ||
Descendants T1 (T-L206); T2 (T-PH110) Defining mutations M184/PAGES34/USP9Y+3178, M272, PAGES129, L810, L455, L452, L445 | ||
Haplogroup T-M184, also known as haplogroup T, is a human Y-chromosome DNA haplogroup. The unique-event polymorphism (UEP) that defines this clade is the single nucleotide polymorphism (SNP) known as M184. Other SNPs – M272, PAGES129, L810, L455, L452, and L445 – are considered to be phylogenetically equivalent to M184.
Contents
- Structure
- Overview
- T1 T L206
- T1a M70
- T1a1
- T1a1a L208
- T1a1a1a1b1a1
- T1a1a1a1b1a1a T Y3836
- T2PH110
- Europe
- Middle East and Caucasus
- South Asia
- Central Asia and East Asia
- Ancient DNA from Karsdorf
- Ancient DNA from Ain Ghazal
- Elite endurance runners
- Thomas Jefferson
- Nomenclatural history
- Original research publications
- References
T-M184 is an immediate descendant of haplogroup LT, whose parent clade is haplogroup K. Before 2008, haplogroup T (or T1a/M70) was known as haplogroup K2, a name that has since been reassigned to a sibling clade of haplogroup LT.
Haplogroup T is unusual in that it is both relatively rare and geographically widespread. The clade probably originated around 40,000 years ago T-M184 is found at its highest frequencies among some populations in East Africa and East India, the arrival of the lineage in these geographical regions is due to relatively recent migration. Males with basal T1* are now found mainly in the Eastern Mediterranean basin.
T2 (T-PH110), the most basal primary branch of T-M184, has been found in three very separate geographical regions: the North European Plain; the Kura-Araks Basin of the Caucasus and; Bhutan. None of these regions, however, now appears to feature populations with high frequencies of haplogroup T-M184.
The other primary branch, Haplogroup T-M206 (T1) is far more common than T2 among modern populations in Eurasia and Africa. It appears to have originated somewhere in western Asia, possibly somewhere between north-eastern Anatolia and the Zagros mountains. T1* may have expanded with the Pre-Pottery Neolithic B culture (PPNB).
Most males who now belong to Haplogroup T-M184 are members of T-M70 (T1a) – a primary branch of T-M206. Now most commonly found in North Africa and the Middle East, T-M70 nevertheless appears to have long been present in Europe and to have arrived there with the first farmers. This is supported by the discovery of several members of T1a1 (CTS880) at a 7,000 year old settlement in Karsdorf, Germany. Autosomal analysis of these remains suggest that some were closely related to modern Southwest Asian populations.
Structure
Overview
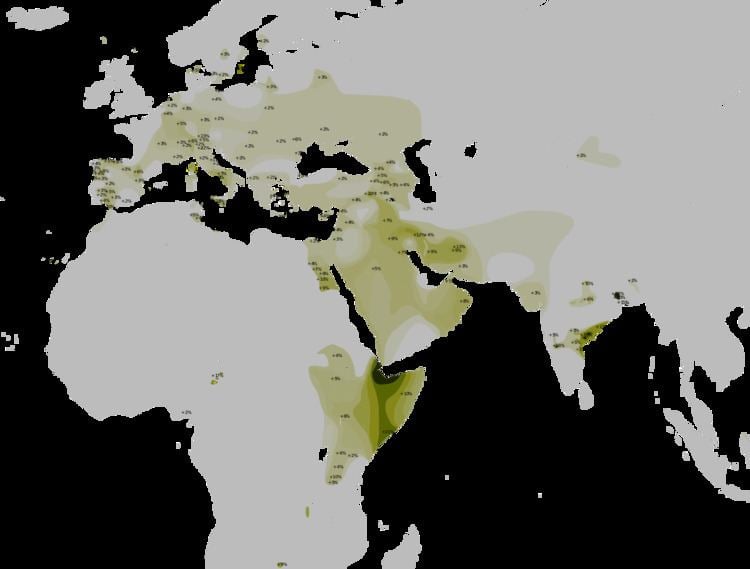
The maximal worldwide frequency for Haplogroup T-M184 is observed in the Dir Clan of Somalia, Djibouti and Ogaden region, where it accounts for approximately 70 to 90% of the male lineages. Luis et al. (2004) suggest that the presence of T on the African continent may, like R1* representatives, point to an older introduction from Asia. The Levant rather than the Arabian Peninsula appears to have been the main route of entry, as the Egyptian and Turkish haplotypes are considerably older in age (13,700 BP and 9,000 BP, respectively) than those found in Oman (only 1,600 BP). According to the authors, the spotty modern distribution pattern of haplogroup T-M184 within Africa may therefore represent the traces of a more widespread early local presence of the clade. Later expansions of populations carrying the E1b1b, E1b1a, G and J NRY lineages may have overwhelmed the T-M184 clade-bearers in certain localities.
In the Caucasus and Anatolia it makes up to 4% of the population in southeast and northwest Caucasus as well as in southeast and western Anatolia, peaking up to 20% in Armenians from Sasun. In Middle East it makes up to 4% of the population around the Zagros Mountains and the Persian Gulf as well as around the Taurus Mountains and the Levant basin, peaking up to 10% in Zoroastrians from Kerman, Bakhtiaris, Assyrians from Azerbaijan, Abudhabians, Armenians from Historical Southwestern Armenia and Druzes from Galilee. In Eastern Africa it makes up to 4% of the population on Upper Egypt and Somalia, peaking up to 10% in Luxor, Jijiga and Dire Dawa.
Haplogroup T is rare almost everywhere in Europe. According to Mendez et al. (2011), the "The occurrence in Europe of lineages belonging to both T1a1 (old T1a) and T1a2 (old T1b) subclades probably reflects multiple episodes of gene flow. T1a1* haplogroups in Europe likely reflect older gene flow ". It makes up to 4% of the population on Central Italy, Western Sicily, Northwest Corsica, Northwestern Iberian Peninsula, Western Andalucia, Western Alps, Eastern Crete, and Macedonia, frequencies up to 10% in Ibiza, Miranda l Douro, Eastern Oviedo, Cádiz, Badajoz, Balagna, Norma and Ragusa, peaking up to 20% in Sciacca, L'Aquila and some German regions. T-M184 was found in 1.7% (10/591) of a pool of six samples of males from southwestern Russia, but it was completely absent from a pool of eight samples totalling 637 individuals from the northern half of European Russia. The Russians from the southwest were from the following cities: Roslavl, Livny, Pristen, Repyevka, and Belgorod; and Kuban Cossacks from the Republic of Adygea.
Besides these regions, T is found in isolated pockets as far as Central Asia, Northeast and Eastern India, Northern Asia, Central Africa, and South Africa. Haplogroup T is found in a majority of Dirs in East Africa, Kurru, Bauris & Lodha in South Asia; and in a significant minority of Rajus and Mahli in South Asia; Somalis, southern Egyptians and Fulbe in north Cameroon; Chian Greeks, Aquilanis, Saccensis, Ibizans (Eivissencs) and Mirandeses in Europe and Zoroastrians, Bakhtiaris in the Middle East.
T1 (T-L206)
T1 is the most common descent of T-M184 haplogroup, being the lineage of more than 95% of all Eurasian T-M184 members. One of their descent lineages is found in high frequencies among northern Somali Clans. However, it appears to have originated somewhere around the northern Mediterranean Basin, perhaps somewhere between Greece to the Zagros mountains.
The basal subclade, T1* appears to have expanded, probably from northeastern Anatolia, into the Levant at least, with the Pre-Pottery Neolithic B culture (PPNB). While it is rare in modern populations, T1* has been found in a Berber individual from Tunisia, a male in Syria and one among ethnic Macedonians in Macedonia.
T1a (M70)
Mendez et al. (2011) point to an ancient presence for T1a-M70 in Europe. The subclade probably arrived with the very first farmers. This is supported by the recent findings of Haak et al. who discovered several T1a1-CTS880 members in a 7000 years old settlement in Karsdorf, Germany.
The T1a1 skeletal remains from this settlement were also found to belong to the H mtdna haplogroup, this settlement has the highest frequency of this mtDNA haplogroup 30.4% (7/23) that have been found in any early Neolithic Europe population until now.
T1a1*
T1a1* (T-L162; T-xL208), which emerged 17,400-14,600 BP, is the largest lineage downstream from T1a-M70 and became widespread across Eurasia and Africa before the modern era.
This extremely rare subclade has been found in Ibizan (Eivissan) islanders and Pontic Greeks from Giresun. The first Y-STR haplotype belonging to this lineage appeared in the paper of Tomas et al in 2006 among a sample of Eivissan individuals but is not until August 2009 when the first T1a1-L162(xL208) individual was reported in a 23andMe customer of Pontic Greek background and Metaxopoulos surname, thanks to the public Adriano Squecco's Y-Chromosome Genome Comparison Project.
Pontic Greeks from Giresun descend from Sinope colonists and Sinope was colonised by Ionians from Miletus. Is interesting to note that there exist an Ionian colony known as Pityussa just like the known Greek name for Eivissa Pityuses. In Eivissa, where is found the famous bust of Demeter that have been confused with the punic Tanit for decades, is known the cult to Demeter. The bust belonging to Demeter have been analysed and is found to contains black particles of volcanic sand origin from the Etna, is thought to be made in Sicily with red clays typical of the eastern Trinacria, which was colonized by the Ionians. The Ionians could be arrived to Eivissa c.2700 YBP. This lineage could be an Ionian marker.
T1a1a (L208)
This lineage, formed 14,200-11,000 BP, is the largest branch downstream T1a1-L162. Firstly discovered and reported at August 2009 in a 23andMe customer of Iberian ancestry that participated in the public Squecco's Y-Chromosome Genome Comparison Project and appearing there as "Avilés" and as "AlpAstur" in 23andMe. Named as "L208" at November 2009.
T1a1a1a1b1a1*
T1a1a1a1b1a1* (T-Y3782*) excluding T1a1a1a1b1a1a (T-Y3836) has been identified only in a single individual from Sardinia.
T1a1a1a1b1a1a (T-Y3836)
This lineage is mostly found among individuals from the Iberian Peninsula, where is found their highest diversity. The first Y-STR haplotype of this lineage, characterized by DYS437=13, was found in the public FTDNA Y-DNA Haplogroup T project, appearing there at April 2009 as kit E8011. However, is not until June 2014 when the Y-SNP Y3836 was discovered in the public YFULL project among two of their participants of Iberian ancestry, appearing there as YF01637 and YF01665.
Actually, two subclades can be clearly discriminated. The first, found mainly in post-colonial Puerto Rico, with DYS391=10 and the second, found mainly in Panamá where their Iberian descendants could have the entrance point to America, with DYS439=12.
Some members of Y3836 are found among different communities of the Sephardic diaspora but they are found to be extremely rare in the total percentage of some of these communities as seen in Nogueiro et al. This probably could mean that these members could be integrated by these communities through the contact with other native Iberian populations as seen in Monteiro et al where this lineage was found among native asturleonese speakers.
T2 (PH110)
This lineage could have arrived in the Levant through the PPNB expansion from northeastern Anatolia.
According to researchers, the frequency in different regions within Germany ranges from 0% to 24% and in the Caucasus from 0% to 12%.
A 2014 study found T-PH110 in one ethnic Bhutanese male, out of a sample of 21, possibly implying a rate of 4.8% in Bhutan.
Europe
With K-M9+, unconfirmed but probable T-M70+ : 14% (3/23) of Russians in Yaroslavl, 12.5% (3/24) of Italians in Matera, 10.3% (3/29) of Italians in Avezzano, 10% (3/30) of Tyroleans in Nonstal, 10% (2/20) of Italians in Pescara, 8.7% (4/46) of Italians in Benevento, 7.8% (4/51) of Italians in South Latium, 7.4% (2/27) of Italians in Paola, 7.3% (11/150) of Italians in Central-South Italy, 7.1% (8/113) of Serbs in Serbia, 4.7% (2/42) of Aromanians in Romania, 3.7% (3/82) of Italians in Biella, 3.7% (1/27) of Andalusians in Córdoba, 3.3% (2/60) of Leoneses in León, 3.2% (1/31) of Italians in Postua, 3.2% (1/31) of Italians in Cavaglià, 3.1% (3/97) of Calabrians in Reggio Calabria, 2.8% (1/36) of Russians in Ryazan Oblast, 2.8% (2/72) of Italians in South Apulia, 2.7% (1/37) of Calabrians in Cosenza, 2.6% (3/114) of Serbs in Belgrade, 2.5% (1/40) of Russians in Pskov, 2.4% (1/42) of Russians in Kaluga, 2.2% (2/89) of Transylvanians in Miercurea Ciuc, 2.2% (2/92) of Italians in Trino Vercellese, 1.9% (2/104) of Italians in Brescia, 1.9% (2/104) of Romanians in Romania, 1.7% (4/237) of Serbs and Montenegrins in Serbia and Montenegro, 1.7% (1/59) of Italians in Marche, 1.7% (1/59) of Calabrians in Catanzaro, 1.6% (3/183) of Greeks in Northern Greece, 1.3% (2/150) of Swiss Germans in Zürich Area, 1.3% (1/79) of Italians in South Tuscany and North Latium, 1.1% (1/92) of Dutch in Leiden, 0.5% (1/185) of Serbs in Novi Sad (Vojvodina), 0.5% (1/186) of Polish in Podlasie
Other parts that have been found to contain a significant proportion of haplogroup T-M184 individuals include Trentino (2/67 or 3%), Mariña Lucense (1/34 or 2.9%), Heraklion (3/104 or 2.9%), Roslavl (3/107 or 2.8%), Ourense (1/37 or 2.7%), Livny (3/110 or 2.7%), Biella (3/114 or 2.6%), Entre Douro (6/228 or 2.6%), Porto (3/118 or 2.5%), Urbino (1/40 or 2.5%), Iberian Peninsula (16/629 or 2.5%), Blekinge/Kristianstad (1/41 or 2.4%), Belarus (1/41 or 2.4%), Modena (3/130 or 2.3%), Provence-Alpes-Côte d'Azur (1/45 or 2.2%), Pristen (1/45 or 2.2%), Cáceres (2/91 or 2.2%), Brac (1/47 or 2.1%), Satakunta (1/48 or 2.1%), Western Croatia (2/101 or 2%), Ukrainia (1/50 or 2%), Greifswald (2/104 or 1.9%), Moldavians in Sofia (1/54 or 1.9%), Uppsala (1/55 or 1.8%), Lublin (2/112 or 1.8%), Pias in Beja (1/54 or 1.8%), Macedonian Greeks (1/57 or 1.8%), Nea Nikomedeia (1/57 or 1.8%), Sesklo/Dimini (1/57 or 1.8%), Lerna/Franchthi (1/57 or 1.8%), Açores (2/121 or 1.7%), Viana do Castelo (1/59 or 1.7%), Toulouse (1/67 or 1.5%), Belgorod (2/143 or 1.4%), Sardinia (1/77 or 1.3%). According to data from commercial testing, 3.9% of Italian males belonging to this haplogroup. Approximately 3% of Sephardi Jews and 2% of Ashkenazi Jews belong to haplogroup T.
Middle East and Caucasus
Haplogroup T has some significant frequencies in southeast and eastern Anatolia, the Zagros Mountains and both sides of the Persian Gulf. Out of 867 recorded by the FTDNA Haplogroup T Project, 284 (32%) are from this area, almost 50% of those from eastern Saudi Arabia.
Unconfirmed but probable T-M70+ : 28% (7/25) of Lezginians in Dagestan, 21.7% (5/23) of Ossetians in Zamankul, 14% (7/50) of Iranians in Isfahan, 13% (3/23) of Ossetians in Zil'ga, 12.6% (11/87) of Kurmanji Kurds in Eastern Turkey, 11.8% (2/17) of Palestinian Arabs in Palestine, 8.3% (1/12) of Iranians in Shiraz, 8.3% (2/24) of Ossetians in Alagir, 8% (2/25) of Kurmanji Kurds in Georgia, 7.5% (6/80) of Iranians in Tehran, 7.4% (10/135) of Palestinian Arabs in Israeli Village, 7% (10/143) of Palestinian Arabs in Israel and Palestine, 5% (1/19) of Chechens in Chechenia, 4.2% (3/72) of Azerbaijanians in Azerbaijan, 4.1% (2/48) of Iranians in Isfahan, 4% (4/100) of Armenians in Armenia, 4% (1/24) of Bedouins in Israel and 2.6% (1/39) of Turks in Ankara.
South Asia
Haplogroup T-M184 has been detected at very high levels in some parts of eastern India.
T1a-M70 in India has been considered to be of West Eurasian origin.
With K-M9+, unconfirmed but probable T-M70+ : 56.6% (30/53) of Kunabhis in Uttar Kannada, 32.5% (13/40) of Kammas in Andhra Pradesh, 26.8% (11/41) of Brahmins in Visakhapatnam, 25% (1/4) of Kattunaiken in South India, 22.4% (11/49) of Telugus in Andhra Pradesh, 20% (1/5) of Ansari in South Asia, (2/20) of Poroja in Andhra Pradesh, 9.8% (5/51) of Kashmiri Pandits in Kashmir, 8.2% (4/49) of Gujars in Kashmir, 7.7% (1/13) of Siddis (migrants from Ethiopia) in Andhra Pradesh, 5.5% (3/55) of Adi in Northeast India, 5.5% (7/128) of Pardhans in Adilabad, 5.3% (2/38) of Brahmins in Bihar, 4.3% (1/23) of Bagata in Andhra Pradesh, 4.2% (1/24) of Valmiki in Andhra Pradesh, (1/32) of Brahmins in Maharashtra, 3.1% (2/64) of Brahmins in Gujarat, 2.9% (1/35) of Rajput in Uttar Pradesh, 2.3% (1/44) of Brahmins in Peruru, and 1.7% (1/59) of Manghi in Maharashtra.
Also in Desasth-Brahmins in Maharashtra (1/19 or 5.3%) and Chitpavan-Brahmins in Konkan (1/21 or 4.8%), Chitpavan-Brahmins in Konkan (2/66 or 3%).
Central Asia and East Asia
Unconfirmed but probable T-M70+ : 2% (4/204) of Hui in Liaoning province, and 0.9% (1/113) of Bidayuh in Sarawak.
Ancient DNA from Karsdorf
Haplogroup T-PF5604, an as-yet unnamed subclade of T1 (upstream from T1a), has been found in the remains of two males who lived 7500–6800 BP, at Karsdorf, Sachsen-Anhalt, Germany. Both T1a skeletal remains belong to the Linienbandkeramische Kultur (LBK). T1a from Karsdorf constitutes 22.2% of all ancient samples between 7500 and 6800 ybp in Germany. The remainder belong to other clades: 22.2% are H2 carriers from Derenburg, and the remaining 55.6% are G2a bearers from Halberstadt and Derenburg. These ancient specimens' mtDNA haplogroups have been found to be H1*/H1au1b and H46b. Their autosomal ancestral components also consist of around 70% Western European Hunter-Gatherer (WHG) and 30% Basal Eurasian.
According to strontium isotope analysis, there are two distinct groups of individuals in Karsdorf but neither were exotic; there was no indication of individuals who grew up in geologically distinct uplands or further north in central Germany. The first group, composed of the majority of the males, could grew up in households that cultivated plots on calcareous soils, very probably in the Unstrut valley in the near vicinity of the settlement. The second group, composed of most of the females, could grew up in households that predominantly cultivated plots on loess, possibly beyond the landmarks of the Unstrut River or about 80m above the site on the Querfurt plateau 1–2 km away. Sex-specific tendencies, the combination of the Sr isotope data with the results of previous carbon and nitrogen isotope analyses, and the similarity of the Sr isotope data of the youngest child with the majority of the males may be evaluated as being in agreement with the predominance of patrilocal residential rules.
In 2015 a published study by Mathieson et al. test several individuals from two Neolithic sites in northwest Anatolia, the results showed that Haplogroup T1a-M70, previously found in LBK sites from Germany, was not present in Barcin nor Mentese Neolithic settlements. This fact together with the absence of the mtDNA lineages carried by both of the T1a individuals from Karsdorf and the occurrence of G2a and the mtDNA lineages carried by all of these G2a individuals, could mean that the Early European Neolithic T1a-M70 had a different migration pattern and, therefore, a different geographical origin.
The autosomal data of I0797 showed the lowest frequency of Anatolian Neolithic component and the highest frequency of an unknown ancient human population for any studied LBK individual. This reinforces the hypothesis of a possible different geographical origin for this T1a tribe instead of the Greco-Anatolian origin of other human groups found in the LBK like G2a.
By his side, I0795 showed higher autosomal admixture frequencies of surrounding populations like Hunter Gatherer Europeans I2a (West Hunter Gatherers) and Aegean-Anatolian Neolithics G2a and H2. However, I0795 have the highest frequency of shared DNA with Upper Paleolithic Neanderthals from Central Europe found in any Early Neolithic population. Further comparisons show that I0795 has similar frequencies like Oase-1 when compared with Vindija Neanderthals. When I0795 and I0797 are compared to Oase-1, they both share a very high percentage of DNA 34% and 18% respectively and I0795 12% with Ostuni1. This could mean that the T1a1 individuals from Karsdorf were closest to Upper Paleolithic Hunter-Gatherers than to Mesolithic haplogroups.
Ancient DNA from 'Ain Ghazal
Haplogroup T is found among the later Middle Pre-Pottery Neolithic B (MPPNB) inhabitants from the 'Ain Ghazal archaeological site (in modern Jordan). It was not found among the early and middle MPPNB populations. It is thought that the Pre-Pottery Neolithic B population is mostly composed of two different populations: members of early Natufian civilisation and a population resulting from immigration from the north, i.e. north-eastern Anatolia. However, Natufians have been found to belong mostly to the E1b1b1b2 lineage – which is found among 60% of the whole PPNB population and 75% of the 'Ain Ghazal population, being present in all three MPPNB stages. Given the complete absence of T-PF7466 among Natufians and earlier MPPNB stages could mean that haplogroup T arrived later with the northerly influx.
As was previously found in the early Neolithic settlement from Karsdorf (Germany) a subclade of mtDNA R0 was found with Y-DNA T at 'Ain Ghazal.
Later MPPNB populations in the Southern Levant were already witnessing severe changes in climate that would have been exacerbated by large population demands on local resources. Beginning at 8.9 cal ka BP we see a significant decrease in population in highland Jordan, ultimately leading to the complete abandonment of almost all central settlements in this region.
The 9th millennium Pre-Pottery Neolithic B (PPNB) period in the Levant represents a major transformation in prehistoric lifeways from small bands of mobile hunter–gatherers to large settled farming and herding villages in the Mediterranean zone, the process having been initiated some 2–3 millennia earlier.
'Ain Ghazal (" Spring of the Gazelles") is situated in a relatively rich environmental setting immediately adjacent to the Wadi Zarqa, the longest drainage system in highland Jordan. It is located at an elevation of about 720m within the ecotone between the oak-park woodland to the west and the open steppe-desert to the east.
Evidence recovered from the excavations suggests that much of the surrounding countryside was forested and offered the inhabitants a wide variety of economic resources. Arable land is plentifull within the site's immediate environs. These variables are atypical of many major neolithic sites in the Near East, several of which are located in marginal environments. Yet despite its apparent richness, the area of 'Ain Ghazal is climatically and environmentally sensitive because of its proximity throughout the Holocene to the fluctuating steppe-forest border.
The Ain Ghazal settlement first appear in the MPPNB and is split into two MPPNB phases. Phase 1 starts 10300 yBP and ends 9950 yBP, phase 2 ends 9550 yBP.
The estimated population of the MPPNB site from ‘Ain Ghazal is of 259-1,349 individuals with an area of 3.01-4.7 ha. Is argued that at its founding at the commencement of the MPPNB ‘Ain Ghazal was likely 2 ha in size and grew to 5 ha by the end of the MPPNB. At this point in time their estimated population was 600-750 people or 125-150 people per hectare.
Elite endurance runners
Possible patterns between Y-chromosome and elite endurance runners were studied in an attempt to find a genetic explanation to the Ethiopian endurance running success. Given the superiority of East African athletes in international distance running over the past four decades, it has been speculated that they are genetically advantaged. Elite marathon runners from Ethiopia were analysed for K*(xP) which according to the previously published Ethiopian studies is attributable to the haplogroup T
According to further studies, T1a1a* (L208) was found to be proportionately more frequent in the elite marathon runners sample than in the control samples than any other haplogroup, therefore this y-chromosome could play a significant role in determining Ethiopian endurance running success. Haplogroup T1a1a* was found in 14% of the elite marathon runners sample of whom 43% of this sample are from Arsi province. In addition, haplogroup T1a1a* was found in only 4% of the Ethiopian control sample and only 1% of the Arsi province control sample. T1a1a* is positively associated with aspects of endurance running, whereas E1b1b1 (old E3b1) is negatively associated.
Thomas Jefferson
A notable member of the T-M184 haplogroup is American President Thomas Jefferson (most distant known ancestor "MDKA" is Samuel Jefferson, Born 11 October 1607 in Pettistree, Suffolk, England). The Y-chromosomal complement of the Jefferson male line was studied in 1998 in an attempt to resolve the controversy over whether he had fathered the mixed-race children of his slave Sally Hemings. A 1998 DNA study of the Y chromosome in the Jefferson male line found that it matched that of a descendant of Eston Hemings, Sally Hemings' youngest son. This confirmed the body of historical evidence, and most historians believe that Jefferson had a long-term intimate liaison with Hemings for 38 years, and fathered her six children of record, four of whom lived to adulthood. In addition, the testing conclusively disproved any connection between the Hemings descendant and the Carr male line. Jefferson grandchildren had asserted in the 19th century that a Carr nephew had been the father of Hemings' children, and this had been the basis of historians' denial for 180 years. Jefferson's paternal family traced back Wales, where T is incredibly rare, as it is throughout Britain. A couple of British males with the Jefferson surname have been found with the third president's type of T, reinforcing the idea that his immediate paternal ancestry was British.
Nomenclatural history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
Original research publications
The following research teams per their publications were represented in the creation of the YCC Tree.
α Jobling and Tyler-Smith 2000 and Kaladjieva 2001
β Underhill 2000
γ Hammer 2001
δ Karafet 2001
ε Semino 2000
ζ Su 1999
η Capelli 2001
