Defining mutations P256 | ||
 | ||
Possible time of origin 32,000-47,000 years BP (Scheinfeldt 2006) Possible place of origin | ||
Haplogroup M, also known as M-P256 and Haplogroup K2b1d is a Y-chromosome DNA haplogroup. M-P256 is a descendant haplogroup of Haplogroup K2b1, and is believed to have first appeared between 32,000 and 47,000 years ago (Scheinfeldt 2006).
Contents
- Phylogenetic structure
- M M P256
- M1 M M4
- M1a M P34
- M1b M P87
- M2 M M353
- M3 M P117
- Previous phylogenetic history
- References
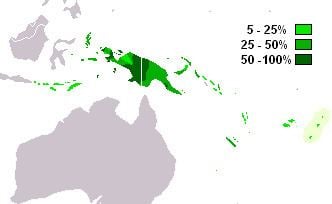
M-P256 is the most frequently occurring Y-chromosome haplogroup in West Papua and western Papua New Guinea.(Kayser 2003). It is also found in neighbouring parts of Melanesia, Indonesia and among indigenous Australians.
Phylogenetic structure
This phylogenetic tree of haplogroup subclades is based primarily on the trees published by ISOGG in 2016 (ISOGG 2016) and YCC in 2008.(Karafet 2008)
M* (M-P256*)
The paragroup M-P256* is found at low incidences in New Guinea (6.3%) and Flores (2.5%).
M1 (M-M4)
Found frequently in New Guinea and Melanesia, with a moderate distribution in neighboring parts of Indonesia, Micronesia, and Polynesia.
An extreme geographical outlier was apparently identified in a 2012 study, which reported a Hazara individual from Mazar-e Sharif, Afghanistan, with M1 among a sample of 60 males from Mazar-e Sharif.(Haber 2012). The Hazara individual carried the SNP M186 (which is believed to be equivalent to M4).
M1a (M-P34)
M1a (M-P34) is the most frequently occurring Y-chromosome DNA haplogroup in Western New Guinea. It is also found with moderate frequency in neighboring parts of Indonesia (Maluku, Nusa Tenggara) and throughout Papua New Guinea, including offshore islands (Karafet 2005 and Kayser 2008).
M1b (M-P87)
M1b M-P87(xM104/P22) has been found in approximately 18% (20/109) of a pool of samples from New Ireland, approximately 12% (5/43) of a sample of Lavongai from New Hanover, approximately 5% (19/395) of a pool of samples from New Britain (and, in particular, in about 24% (15/63) of Baining from East New Britain), in one Saposa individual from northern Bougainville, and in another individual from the north coast of Papua New Guinea (Scheinfeldt 2006).
The subclade M1b1 (M104_1/P22_1, M104_2/P22_2) is found frequently in populations of the Bismarck Archipelago and Bougainville Island, with a moderate distribution in New Guinea, Fiji, Tonga, East Futuna, and Samoa. (Kayser 2008 and Scheinfeldt 2006).
M2 (M-M353)
Found at a low frequency in Fiji and East Futuna (Kayser 2006).
The subclade M2a (M-M177 a.k.a. M-SRY9138) is found in one Nasioi individual from the eastern coast of Bougainville and in one individual from Malaita Province of the Solomon Islands (Cox 2006).
Historic names for M-SRY9138 (a.k.a. M-M177) from peer reviewed literature.
M3 (M-P117)
M3 (P117, P118) is found frequently in populations of New Britain, and also observed occasionally in northern Bougainville, Fiji, and East Futuna (Kayser 2008 and Scheinfeldt 2006).
Previous phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
The following research teams per their publications were represented in the creation of the YCC Tree.
Karafet's 2008 paper introduced a number of changes, compared to the previous 2006 ISOGG tree. Before the discovery of the P256 marker, the current subgroup M-M4 (defined by the M4 marker) previously represented the whole of Haplogroup M-P256; and subgroups M2 and M3 were formerly classed as subgroups K1 and K7 of the parent Haplogroup K.
