Ancestor Haplogroup R1 | ||
 | ||
Possible time of origin 22,000 YBP to 25,000 years ago Descendants Haplogroup R1a-Z282 (Europe), R1a-Z93 (Asia) Defining mutations R1a: L62, L63, L120, M420, M449, M511, M513R1a1a: M17, M198, M512, M514, M515, L168, L449, L457, L566 | ||
Haplogroup R1a, or haplogroup R-M420, is a human Y-chromosome DNA haplogroup which is distributed in a large region in Eurasia, extending from Scandinavia, Central Europe and southern Siberia to South Asia.
Contents
- R1a origins
- Diversification of R1a1a1 M417 and ancient migrations
- Steppe origins
- Transcaucasia West Asian origins and possible influence on Indus Valley Civilisation
- Proposed South Asian origins
- Phylogeny
- Topology
- R M173 R1
- R M420 R1a
- R SRY15322 R1a1
- R M17M198 R1a1a
- R M17 R1a1a1
- R Z282 R1a1a1b1a Eastern Europe
- R1a1a1b2 R Z93 Asia
- Historical
- Europe
- Central Asia
- South Asia
- East Asia
- West Asia
- Popular science
- In art
- Historic naming of R1a
- References
While R1a originated ca. 22,000 to 25,000 years ago, its subclade M417 (R1a1a1) diversified ca. 5,800 years ago. The distribution of M417-subclades R1-Z282 (including R1-Z280) in Central- and Eastern Europe and R1-Z93 in Asia suggests that R1a1a diversified within the Eurasian Steppes or the Middle East and Caucasus region. The place of origin of these subclades plays a role in the debate about the origins of Indo-Europeans.
The SNP mutation R-M420 was discovered after R-M17 (R1a1a), which resulted in a reorganization of the lineage in particular establishing a new paragroup (designated R-M420*) for the relatively rare lineages which are not in the R-SRY10831.2 (R1a1) branch leading to R-M17.
R1a origins
The split of R1a (M420) is computed to ca. 22,000, or 25,000 years ago, which is the time of the last glacial maximum. A large, 2014 study by Peter A Underhill et al., using 16,244 individuals from over 126 populations from across Eurasia, concluded that there was compelling evidence that "the initial episodes of haplogroup R1a diversification likely occurred in the vicinity of present-day Iran."
Diversification of R1a1a1 (M417) and ancient migrations
According to Underhill (2014), the downstream R1a-M417 subclade diversified into Z282 and Z93 circa 5,800 years ago. The question of the origins of R1a1a is relevant to the ongoing debate concerning the urheimat of the proto-Indo-European people, and may also be relevant to the origins of the Indus Valley Civilisation. R1a shows a strong correlation with Indo-European languages of western/southern Asia and eastern Europe, being most prevalent in Poland, Russia, and Ukraine, and in central Asia, Afghanistan, Pakistan and India. In Eastern Europe Z282 is prevalent, while in South Asia Z93 dominates. The connection between Y-DNA R-M17 and the spread of Indo-European languages was first noted by T. Zerjal and colleagues in 1999.
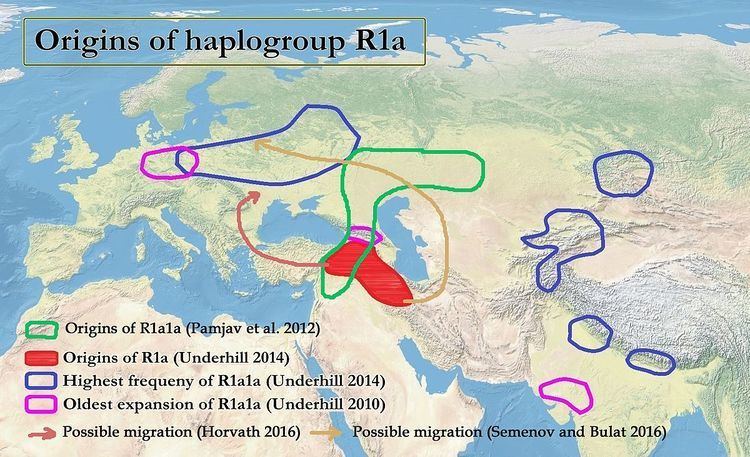
Steppe origins
Proposed steppe dispersal of R1a1a
Kivisild et al. (2003) have proposed either south or west Asia, while Mirabal et al. (2009) see support for both south and central Asia. Other studies suggest Ukrainian, Central Asian and West Asian origins for R1a1a.
Ornella Semino et al. (2000) proposed Ukrainian origins, and a postglacial spread of the R1a1 gene during the Late Glacial Maximum, subsequently magnified by the expansion of the Kurgan culture into Europe and eastward. Spencer Wells proposes central Asian origins, suggesting that the distribution and age of R1a1 points to an ancient migration corresponding to the spread by the Kurgan people in their expansion from the Eurasian steppe. According to Pamjav et al. (2012), R1a1a diversified in the Eurasian Steppes or the Middle East and Caucasus region:
Inner and Central Asia is an overlap zone for the R1a1-Z280 and R1a1-Z93 lineages [which] implies that an early differentiation zone of R1a1-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Central- and Eastern Europe."
Source of R1a1a1 in Corded Ware culture
David Anthony considers the Yamna culture to be the Indo-European Urheimat. According to Haak et al. (2015), a massive migration from the Yamna culture northwards took place ca. 2,500 BCE, accounting for 75% of the genetic ancestry of the Corded Ware culture, noting that R1a and R1b may have "spread into Europe from the East after 3,000 BCE". Yet, all their seven Yamna samples belonged to the R1b-M269 subclade, but no R1a1a has been found in their Yamna samples. This raises the question where the R1a1a in the Corded Ware culture came from, if it was not from the Yamna culture.
R1a may have migrated from the Anatolian-Iranian area via Central Asia to Eastern Europe, in concreto the Comb Ware culture (4,200 BCE – 2,000 BCE), which was partly absorbed by the Corded Ware culture. R1a1 has been found in samples from the Narva culture, which was part of the Comb Ware culture. Horvath rejects this possible migration route, given the dominance of haplogroup N1c in the Comb Ware culture, and that the Corded ware autosomal DNA is derived from the Yamna culture, and not from the Comb Ware culture. In contrast, Semenov and Bulat do argue for such an origin of R1a1a in the Corded ware culture, noting that several publications point to the presence of R1a1 in the Comb Ware culture.
Horvath proposes a migration of R1a from the Anatolian-Iranian area to the Pontic steppe via the Balkan. Horvath notes that Haak et al. (2015) found that part of the Yamna ancestry derived from the Middle East, and that neolithic techniques probably arrived at the Yamna culture from the Balkans. Horvath further notes that in the area of the Rossen culture (4,600–4,300 BC), which was situated on Germany and predates the Corded Ware culture, an old subclade of R1a, namely L664, can still be found. From these facts Horvath speculates that R1a arrived in the Balkans via Anatolia, and from there spread first north-west to the Rossen culture, and then east from the Cucuteni culture to the Yamna and Afanasevo cultures, despite the absence of R1a from intermediate cultures between the Near East, Anatolia and the Balkans.
Transcaucasia & West Asian origins and possible influence on Indus Valley Civilisation
Part of the Indian genetic ancestry derives from west Eurasian populations, and some researchers have implied that Z93 may have come to India via Iran and expanded there during the Indus Valley Civilisation.
Mascarenhas et al. (2015) note that the roots of Z93 lie in West Asia, and propose that "Z93 and L342.2 expanded in a southeasterly direction from Transcaucasia into South Asia," noting that such an expansion is compatible with "the archeological records of eastward expansion of West Asian populations in the 4th millennium BCE culminating in the so-called Kura-Araxes migrations in the post-Uruk IV period." Yet, Lazaridis noted that sample I1635 of Lazaridis et al. (2016), their Armenian Kura-Araxes sample, carried Y-haplogroup R1b1-M415(xM269) (also called R1b1a1b-CTS3187).
According to Underhill et al. (2014/2015) the diversification of Z93 and the "early urbanization within the Indus Valley [...] occurred at [5,600 years ago] and the geographic distribution of R1a-M780 (Figure 3d) may reflect this." Poznik et al. (2016) note that 'striking expansions' occurred within R1a-Z93 at ~4,500–4,000 years ago, which "predates by a few centuries the collapse of the Indus Valley Civilisation."
Proposed South Asian origins
Kivisild et al. (2003) have proposed either south or west Asia, while Mirabal et al. (2009) see support for both south and central Asia.
South Asian populations have the highest STR diversity within R1a1a, and subsequent older TMRCA datings, and R1a1a is present among both higher (Brahmin) castes and lower castes, although the presence is substantially higher among Brahmin castes. From these findings some researchers have concluded that R1a1a originated in south Asia, excluding a substantial genetic influx from Indo-European migrants.
Yet, this diversity can also be explained by the historically high population numbers, which increases the likelihood of diversification and microsatellite variation. The idea of Indian origins of R1a1 also implies a migration of Indo-European genes and languages "Out of India" to Europe and east Asia. This is incompatible with the mainstream scholarly view, which states that the proto-Indo-Aryan language originated outside India. And according to Sengupta et al. (2006), "[R1a1 and R2] could have actually arrived in southern India from a southwestern Asian source region multiple times."
Phylogeny
The R1a family tree now has three major levels of branching, with the largest number of defined subclades within the dominant and best known branch, R1a1a (which will be found with various names; in particular, as "R1a1" in relatively recent but not the latest literature.)
Topology
The topology of R1a is as follows (codes [in brackets] non-isogg codes): Tatiana et al. (2014) "rapid diversification process of K-M526 likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q."
R-M173 (R1)
R1a is distinguished by several unique markers, including the M420 mutation. It is a subclade of Haplogroup R-M173 (previously called R1). R1a has the sister-subclades Haplogroup R1b-M343, and the paragroup R-M173*.
R-M420 (R1a)
R-M420, defined by the mutation M420, has two branches: R-SRY1532.2, defined by the mutation SRY1532.2, which makes up the vast majority; and R-M420*, the paragroup, defined as M420 positive but SRY1532.2 negative. (In the 2002 scheme, this SRY1532.2 negative minority was one part of the relatively rare group classified as the paragroup R1*.) Mutations understood to be equivalent to M420 include M449, M511, M513, L62, and L63.
Only isolated samples of the new paragroup R-M420* were found by Underhill 2009, mostly in the Middle East and Caucasus: 1/121 Omanis, 2/150 Iranians, 1/164 in the United Arab Emirates, and 3/612 in Turkey. Testing of 7224 more males in 73 other Eurasian populations showed no sign of this category.
R-SRY1532.2 (R1a1)
R1a1 is defined by SRY1532.2 or SRY10831.2), understood to always include SRY10831.2, M448, L122, M459, and M516.) This family of lineages is dominated by M17 and M198. In contrast, paragroup R-SRY1532.2* lacks either the M17 or M198 markers.
The R-SRY1532.2* paragroup is apparently less rare than R1*, but still relatively unusual, though it has been tested in more than one survey. Underhill et all. (2009) reported 1/51 in Norway, 3/305 in Sweden, 1/57 Greek Macedonians, 1/150 Iranians, 2/734 ethnic Armenians, and 1/141 Kabardians. Sahoo et al. (2006) reported R-SRY1532.2* for 1/15 Himachal Pradesh Rajput samples.
R-M17/M198 (R1a1a)
The following SNPs are associated with R1a1a:
R-M17 (R1a1a1)
R1a1a1 (RM-417) is the most widely found subclade, in two variations which are found respectively in Europe (R1a1a1b1 (R-Z282) ([R1a1a1a*] (R-Z282) (Underhill 2014/2015)) and Central and South Asia (R1a1a1b2 (R-Z93) ([R1a1a2*] (R-Z93) Underhill 2014/2015)).
R-Z282 (R1a1a1b1a) (Eastern Europe)
This large subclade appears to encompass most of the R1a1a found in Europe.
R-M458 (R1a1a1b1a1)
R-M458 is a mainly Slavic SNP, characterized by its own mutation, and was first called cluster N. Underhill et al. (2009) found it to be present in modern European populations roughly between the Rhine catchment and the Ural Mountains and traced it to "a founder effect that [...] falls into the early Holocene period, 7.9±2.6 KYA." M458 was found in one skeleton from a 14th-century grave field in Usedom, Mecklenburg-Vorpommern, Germany. The paper by Underhill et al. (2009) also reports a surprisingly high frequency of M458 in some Northern Caucasian populations (for example 27.5% among Karachays and 23.5% among Balkars, 7.8% among Karanogays and 3.4% among Abazas).
R-L260 (R1a1a1b1a1a) (Gwozdz's cluster P)
R1a1a1b1a1a (R-L260), commonly referred to as West Slavic or Polish, is a subclade of the larger parent group R-M458, and was first identified as an STR cluster by Pawlowski 2002 and then by Gwozdz 2009. Thus, R-L260 was what Gwozdz 2009 called cluster "P." In 2010 it was verified to be a haplogroup identified by its own mutation (SNP). It apparently accounts for about 8% of Polish men, making it the most common subclade in Poland. Outside of Poland it is less common (Pawlowski 2002). In addition to Poland, it is mainly found in the Czech Republic and Slovakia, and is considered "clearly West Slavic." The founding ancestor of R-L260 is estimated to have lived between 2000 and 3000 years ago, i.e. during the Iron Age, with significant population expansion less than 1,500 years ago.
R-M334
R-M334 ([R1a1a1g1], a subclade of [R1a1a1g] (M458) c.q. R1a1a1b1a1 (M458)) was found by Underhill et al. (2009) only in one Estonian man and may define a very recently founded and small clade.
R1a1a1b1a2 (S466/Z280, S204/Z91)
R1a1a1b1a2b3* (Gwozdz's Cluster K)
R1a1a1b1a2b3* (M417+, Z645+, Z283+, Z282+, Z280+, CTS1211+, CTS3402, Y33+, CTS3318+, Y2613+) (Gwozdz's Cluster K) is a STR based group that is R-M17(xM458). This cluster is common in Poland but not exclusive to Poland.
R1a1a1b1a2b3a (R-L365)
R1a1a1b1a2b3a (R-L365) was early called Cluster G.
R1a1a1b2 (R-Z93) (Asia)
This large subclade appears to encompass most of the R1a1a found in Asia.
Historical
Haplogroup R1a has been found in ancient fossils associated with the Corded Ware culture and Urnfield culture; as well as the burial of the remains of the Sintashta culture, Andronovo culture, the Pazyryk culture, Tagar culture and Tashtyk culture, the inhabitants of ancient Tanais, in the Tarim mummies, the aristocracy Xiongnu. in two ancient Khazar fossils. The skeletal remains of a father and his two sons, from an archaeological site discovered in 2005 near Eulau (in Saxony-Anhalt, Germany) and dated to about 2600 BCE, tested positive for the Y-SNP marker SRY10831.2. The Ysearch number for the Eulau remains is 2C46S. The ancestral clade was thus present in Europe at least 4600 years ago, in association with one site of the widespread Corded Ware culture.
Europe
In Europe, the R1a1 sub-clade is found at highest levels among peoples of Eastern European descent, with 50 to 65% among Sorbs, Poles, Russians and Ukrainians. In the Baltic countries R1a1a frequencies decrease from Lithuania (45%) to Estonia (around 30%). Levels in Hungarians have been noted between 20 and 60%.
There is a significant presence in peoples of Scandinavian descent, with highest levels in Norway and Iceland, where between 20 and 30% of men are in R1a1a. Vikings and Normans may have also carried the R1a1a lineage westward; accounting for at least part of the small presence in the British Isles. In East Germany, where Haplogroup R1a1a reaches a peak frequency in Rostock at a percentage of 31.3%, it averages between 20 and 30%.
Haplogroup R1a1a was found at elevated levels among a sample of the Israeli population who self-designated themselves as Levites and Ashkenazi Jews (Levites comprise approximately 4% of Jews). Behar reported R1a1a to be the dominant haplogroup in Ashkenazi Levites (52%), although rare in Ashkenazi Cohanim (1.3%).
In Southern Europe R1a1a is not common, but significant levels have been found in pockets, such as in the Pas Valley in Northern Spain, areas of Venice, and Calabria in Italy. The Balkans shows lower frequencies, and significant variation between areas, for example more than 30% in Slovenia, Croatia and Greek Macedonia, but less than 10% in Albania, Kosovo and parts of Greece.
Central Asia
In Afghanistan, R1a1a is found at 51% among the Pashtuns who are the largest ethnic group in Afghanistan, 50% among the Kyrgyz, and 30% among the Tajiks. It is less frequent among the Hazaras (7%) and the Turkic-speaking Uzbeks (18%).
South Asia
In South Asia, R1a1a has often been observed with high frequency in a number of demographic groups.
In India, high frequencies of this haplogroup is observed in West Bengal Brahmins (72%) to the east, Konkanastha Brahmins (48%) to the west, Khatris (67%) in the north and Iyengar Brahmins (31%) in the south. It has also been found in several South Indian Dravidian-speaking Adivasis including the Chenchu (26%) and the Valmikis of Andhra Pradesh and the Kallar of Tamil Nadu suggesting that R1a1a is widespread in Tribal Southern Indians.
Besides these, studies show high percentages in regionally diverse groups such as Manipuris (50%) to the extreme North East and in Punjab (47%) to the extreme North West.
In Pakistan it is found at 71% among the Mohanna tribe in Sindh province to the south and 46% among the Baltis of Gilgit-Baltistan to the north. Among the Sinhalese of Sri Lanka, 13% were found to be R1a1a (R-SRY1532) positive in a sample size of 39 subjects. Hindus of Terai region of Nepal show it at 69%.
East Asia
The frequency of R1a1a is comparatively low among some Turkic-speaking groups including Turks, Azeris, Kazakhs, and Yakuts, yet levels are higher (19 to 28%) in certain Turkic or Mongolic-speaking groups of Northwestern China, such as the Bonan, Dongxiang, Salar, and Uyghurs.
In Eastern Siberia, R1a1a is found among certain indigenous ethnic groups including Kamchatkans and Chukotkans, and peaking in Itel'man at 22%.
West Asia
R1a1a has been found in various forms, in most parts of Western Asia, in widely varying concentrations, from almost no presence in areas such as Jordan, to much higher levels in parts of Kuwait, Turkey and Iran. The Shimar (Shammar) Bedouin tribe in Kuwait show the highest frequency in the Middle East at 43%.)
Wells 2001, noted that in the western part of the country, Iranians show low R1a1a levels, while males of eastern parts of Iran carried up to 35% R1a1a. Nasidze 2004 found R1a1a in approximately 20% of Iranian males from the cities of Tehran and Isfahan. Regueiro 2006 in a study of Iran, noted much higher frequencies in the south than the north.
A newer Study has found 20.3% R-M17* among Kurdish samples which were taken in the Kurdistan Province in western Iran, 9.7% among Mazandaranis in North Iran in the province of Mazandaran, 9.4% among Gilaks in province of Gilan, 12.8% among Persian and 17.6% among Zoroastrians in Yazd, 18.2% among Persians in Isfahan, 20.3% among Persians in Khorasan, 16.7% Afro-Iranians, 18.4% Qeshmi "Gheshmi", 21.4% among Persian Speaking Bandari people in Hormozgan and 25% among the Baloch people in Sistan and Baluchestan Province.
Further to the north of these Middle Eastern regions on the other hand, R1a1a levels start to increase in the Caucasus, once again in an uneven way. Several populations studied have shown no sign of R1a1a, while highest levels so far discovered in the region appears to belong to speakers of the Karachay-Balkar language among whom about one quarter of men tested so far are in haplogroup R1a1a.
Popular science
Bryan Sykes in his book Blood of the Isles gives imaginative names to the founders or "clan patriarchs" of major British Y haplogroups, much as he did for mitochondrial haplogroups in his work The Seven Daughters of Eve. He named R1a1a in Europe the "clan" of a "patriarch" Sigurd, reflecting the theory that R1a1a in the British Isles has Norse origins.
In art
Artem Lukichev created an animation based on the Bashkir epic about the Ural, which outlined the history of the clusters of haplogroup R1: R1a and R1b.
Historic naming of "R1a"
The historic naming system commonly used for R1a was inconsistent in different published sources, because it changed often; this requires some explanation.
In 2002, the Y Chromosome Consortium (YCC) proposed a new naming system for haplogroups (YCC 2002), which has now become standard. In this system, names with the format "R1" and "R1a" are "phylogenetic" names, aimed at marking positions in a family tree. Names of SNP mutations can also be used to name clades or haplogroups. For example, as M173 is currently the defining mutation of R1, R1 is also R-M173, a "mutational" clade name. When a new branching in a tree is discovered, some phylogenetic names will change, but by definition all mutational names will remain the same.
The widely occurring haplogroup defined by mutation M17 was known by various names, such as "Eu19", as used in (Semino 2000) in the older naming systems. The 2002 YCC proposal assigned the name R1a to the haplogroup defined by mutation SRY1532.2. This included Eu19 (i.e. R-M17) as a subclade, so Eu19 was named R1a1. Note, SRY1532.2 is also known as SRY10831.2 The discovery of M420 in 2009 has caused a reassignment of these phylogenetic names.(Underhill 2009 and ISOGG 2012) R1a is now defined by the M420 mutation: in this updated tree, the subclade defined by SRY1532.2 has moved from R1a to R1a1, and Eu19 (R-M17) from R1a1 to R1a1a.
More recent updates recorded at the ISOGG reference webpage involve branches of R-M17, including one major branch, R-M417.
