 | ||
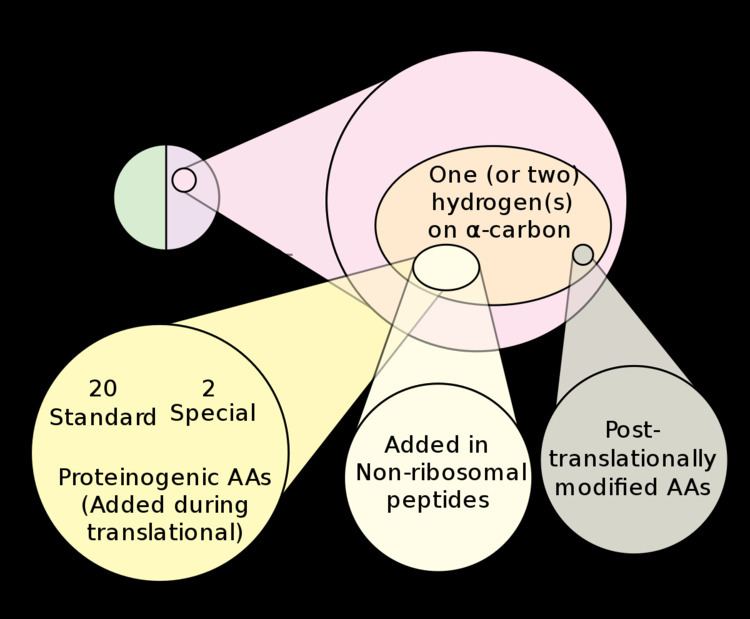
Proteinogenic amino acids are amino acids that are incorporated biosynthetically into proteins during translation. The word "proteinogenic" means "protein creating". Throughout known life, there are 22 genetically encoded (proteinogenic) amino acids, 20 in the standard genetic code and an additional 2 that can be incorporated by special translation mechanisms.
Contents
- Structures
- Chemical properties
- Side chain properties
- Gene expression and biochemistry
- Mass spectrometry
- Stoichiometry and metabolic cost in cell
- Life based on alternative proteinogenic sets
- References
In contrast, non-proteinogenic amino acids are amino acids that are either not incorporated into proteins (like GABA, L-DOPA, or triiodothyronine), misincorporated in place of a genetically encoded amino acid, or not produced directly and in isolation by standard cellular machinery (like hydroxyproline). The latter often results from post-translational modification of proteins. Some non-proteinogenic amino acids are incorporated into nonribosomal peptides which are synthesized by non-ribosomal peptide synthetases.
Both eukaryotes and prokaryotes can incorporate selenocysteine into their proteins via a nucleotide sequence known as a SECIS element, which directs the cell to translate a nearby UGA codon as selenocysteine (UGA is normally a stop codon). In some methanogenic prokaryotes, the UAG codon (normally a stop codon) can also be translated to pyrrolysine.
In eukaryotes, there are only 21 proteinogenic amino acids, the 20 of the standard genetic code, plus selenocysteine. Humans can synthesize 12 of these from each other or from other molecules of intermediary metabolism. The other nine must be consumed (usually as their protein derivatives), and so they are called essential amino acids. The essential amino acids are histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine (i.e. H, I, L, K, M, F, T, W, V).
The proteinogenic amino acids have been found to be related to the set of amino acids that can be recognized by ribozyme autoaminoacylation systems. Thus, non-proteinogenic amino acids would have been excluded by the contingent evolutionary success of nucleotide-based life forms. Other reasons have been offered to explain why certain specific non-proteinogenic amino acids are not generally incorporated into proteins; for example, ornithine and homoserine cyclize against the peptide backbone and fragment the protein with relatively short half-lives, while others are toxic because they can be mistakenly incorporated into proteins, such as the arginine analog canavanine.
Structures
The following illustrates the structures and abbreviations of the 21 amino acids that are directly encoded for protein synthesis by the genetic code of eukaryotes. The structures given below are standard chemical structures, not the typical zwitterion forms that exist in aqueous solutions.
IUPAC/IUBMB now also recommends standard abbreviations for the following two amino acids:
Chemical properties
Following is a table listing the one-letter symbols, the three-letter symbols, and the chemical properties of the side chains of the standard amino acids. The masses listed are based on weighted averages of the elemental isotopes at their natural abundances. Forming a peptide bond results in elimination of a molecule of water, so the mass of an amino acid unit within a protein chain is reduced by 18.01524 Da.
General chemical properties
Side chain properties
§: Values for Asp, Cys, Glu, His, Lys & Tyr were determined using the amino acid residue placed centrally in an alanine pentapeptide. The value for Arg is from Pace et al. (2009). The value for Sec is from Byun & Kang (2011).
N.D.: The pKa value of Pyrrolysine has not been reported.
Note: The pKa value of an amino-acid residue in a small peptide is typically slightly different when it is inside a protein. Protein pKa calculations are sometimes used to calculate the change in the pKa value of an amino-acid residue in this situation.
Gene expression and biochemistry
* UAG is normally the amber stop codon, but encodes pyrrolysine if a PYLIS element is present.
** UGA is normally the opal (or umber) stop codon, but encodes selenocysteine if a SECIS element is present.
† The stop codon is not an amino acid, but is included for completeness.
†† UAG and UGA do not always act as stop codons (see above).
‡ An essential amino acid cannot be synthesized in humans and must, therefore, be supplied in the diet. Conditionally essential amino acids are not normally required in the diet, but must be supplied exogenously to specific populations that do not synthesize it in adequate amounts.
& Occurrence of amino acids is based on 135 Archaea, 3775 Bacteria, 614 Eukaryota proteomes and human proteome (21 006 proteins) respectively.
Mass spectrometry
In mass spectrometry of peptides and proteins, knowledge of the masses of the residues is useful. The mass of the peptide or protein is the sum of the residue masses plus the mass of water (Monoisotopic mass = 18.01056 Da; average mass = 18.0153 Da). The residue masses are calculated from the tabulated chemical formulas and atomic weights. In mass spectrometry, ions may also include one or more protons (Monoisotopic mass = 1.00728 Da; average mass = 1.0074 Da).
§ Monoisotopic mass
Stoichiometry and metabolic cost in cell
The table below lists the abundance of amino acids in E.coli cells and the metabolic cost (ATP) for synthesis the amino acids. Negative numbers indicate the metabolic processes are energy favorable and do not cost net ATP of the cell. The abundance of amino acids includes amino acids in free form and in polymerization form (proteins).
Life based on alternative proteinogenic sets
The proteinogenic set used by known life on Earth appears to be arbitrarily selected by evolution, according to current knowledge, from many hundreds of possible alpha-type amino acids. Xenobiology studies hypothetical life forms that could be constructed using alternative sets using expanded genetic codes. Miller-type experiments on artificial abiogenesis show that alpha-type amino acids predominate in water-based 'primordial soups', but beta-type amino acids dominate when less water is present. Both alpha- and beta-based sets could form the basis for alternative protein constructions and life forms.
