Formula C3H7NO2Se | ||
 | ||
What is the definition of selenocysteine medical school terminology dictionary
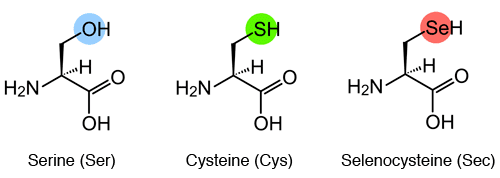
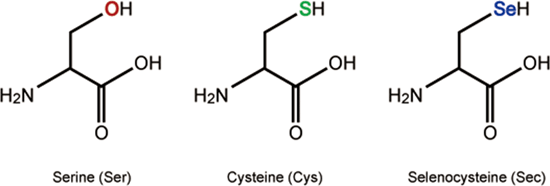
Selenocysteine (abbreviated as Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid.
Contents
- What is the definition of selenocysteine medical school terminology dictionary
- Selenocysteine meaning
- Structure
- Biology
- Applications
- References
Selenocysteine exists naturally in three domains of life, but not in every lineage, as a building block of selenoproteins. Selenocysteine is a cysteine analogue with a selenium-containing selenol group in place of the sulfur-containing thiol group.
Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5' deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 2, methionine-R-sulfoxide reductase B1 (SEPX1), and some hydrogenases).
Selenocysteine was discovered by biochemist Thressa Stadtman at the National Institutes of Health.
Selenocysteine meaning
Structure
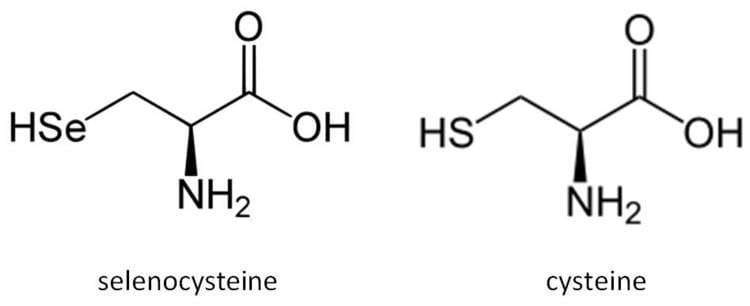
Selenocysteine has a structure similar to that of cysteine, but with an atom of selenium taking the place of the usual sulfur, forming a selenol group which is deprotonated at physiological pH. Proteins that contain one or more selenocysteine residues are called selenoproteins. Most selenoproteins contain a single Sec residue. Selenoproteins with catalytic activities which depend on selenocysteine's biochemical activity are called selenoenzymes. The structurally characterized selenoenzymes have been found to employ catalytic triad structures that influence the nucleophilicity of the active site selenocysteine.
Biology
Selenocysteine has both a lower pKa (5.47) and a lower reduction potential than cysteine. These properties make it very suitable in proteins that are involved in antioxidant activity.
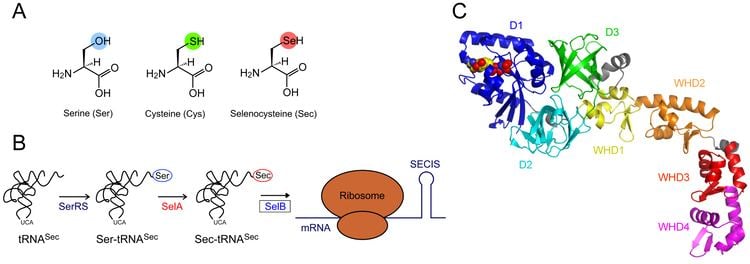
Although it is found in the three domains of life, it is not universal in all organisms. Unlike other amino acids present in biological proteins, selenocysteine is not coded for directly in the genetic code. Instead, it is encoded in a special way by a UGA codon, which is normally a stop codon. Such a mechanism is called translational recoding and its efficiency depends on the selenoprotein being synthesized and on translation initiation factors. When cells are grown in the absence of selenium, translation of selenoproteins terminates at the UGA codon, resulting in a truncated, nonfunctional enzyme. The UGA codon is made to encode selenocysteine by the presence of a selenocysteine insertion sequence (SECIS) in the mRNA. The SECIS element is defined by characteristic nucleotide sequences and secondary structure base-pairing patterns. In bacteria, the SECIS element is typically located immediately following the UGA codon within the reading frame for the selenoprotein. In Archaea and in eukaryotes, the SECIS element is in the 3' untranslated region (3' UTR) of the mRNA, and can direct multiple UGA codons to encode selenocysteine residues.
Again unlike the other amino acids, no free pool of selenocysteine exists in the cell. Its high reactivity would cause damage to cells. Instead, cells store selenium in the less reactive selenide form (H2Se). Selenocysteine synthesis occurs on a specialized tRNA, which also functions to incorporate it into nascent polypeptides.
The primary and secondary structure of selenocysteine-specific tRNA, tRNASec, differ from those of standard tRNAs in several respects, most notably in having an 8-base (bacteria) or 10-base (eukaryotes) pair acceptor stem, a long variable region arm, and substitutions at several well-conserved base positions. The selenocysteine tRNAs are initially charged with serine by seryl-tRNA ligase, but the resulting Ser-tRNASec is not used for translation because it is not recognised by the normal translation elongation factor (EF-Tu in bacteria, eEF1A in eukaryotes).
Rather, the tRNA-bound seryl residue is converted to a selenocysteine residue by the pyridoxal phosphate-containing enzyme selenocysteine synthase. In eukaryotes and archaea two enzymes are required to convert tRNA-bound seryl residue intro tRNA selenocysteinyl residue: PSTK [O-phosphoseryl-tRNA[Ser]Sec kinase]) and selenocysteine synthase. Finally, the resulting Sec-tRNASec is specifically bound to an alternative translational elongation factor (SelB or mSelB (or eEFSec)), which delivers it in a targeted manner to the ribosomes translating mRNAs for selenoproteins. The specificity of this delivery mechanism is brought about by the presence of an extra protein domain (in bacteria, SelB) or an extra subunit (SBP2 for eukaryotic mSelB/eEFSec) which bind to the corresponding RNA secondary structures formed by the SECIS elements in selenoprotein mRNAs.
As of 2016, fifty-four human proteins are known to contain selenocysteine (selenoproteins).
Selenocysteine derivatives γ-glutamyl-Se-methylselenocysteine and Se-methylselenocysteine occur naturally in plants of the genera Allium and Brassica.
Applications
Biotechnological applications of selenocysteine include use of 73Se-labeled Sec (half-life of 73Se = 7.2 hours) in positron emission tomography (PET) studies and 75Se-labeled Sec (half-life of 75Se = 118.5 days) in specific radiolabeling, facilitation of phase determination by multi-wavelength anomalous diffraction in X-ray crystallography of proteins by introducing Sec alone, or Sec together with selenomethionine (SeMet), and incorporation of the stable 77Se isotope, which has a nuclear spin of 1/2 and can be used for high-resolution NMR, among others.
