 | ||
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring. HGT is an important factor in the evolution of many organisms.
Contents
- History
- Mechanism
- Inference
- Viruses
- Prokaryotes
- Bacterial transformation
- Bacterial conjugation
- Archaeal DNA transfer
- Eukaryotes
- Horizontal transposon transfer
- Artificial horizontal gene transfer
- Importance in evolution
- Challenge to the tree of life
- Genes
- References
Horizontal gene transfer is the primary mechanism for the spread of antibiotic resistance in bacteria, plays an important role in the evolution of bacteria that can degrade novel compounds such as human-created pesticides and in the evolution, maintenance, and transmission of virulence. It often involves temperate bacteriophages and plasmids. Genes responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various mechanisms such as F-pilus, subsequently arming the antibiotic resistant genes' recipient against antibiotics, which is becoming a medical challenge to deal with.
Most thinking in genetics has focused upon vertical transfer, but horizontal gene transfer is important, and among single-celled organisms is perhaps the dominant form of genetic transfer.
Artificial horizontal gene transfer is a form of genetic engineering.
History
Horizontal genetic transfer was first described in Seattle in 1951, in a paper demonstrating that the transfer of a viral gene into Corynebacterium diphtheriae created a virulent from a non-virulent strain, also simultaneously solving the riddle of diphtheria (that patients could be infected with the bacteria but not have any symptoms, and then suddenly convert later or never), and giving the first example for the relevance of the lysogenic cycle. Inter-bacterial gene transfer was first described in Japan in a 1959 publication that demonstrated the transfer of antibiotic resistance between different species of bacteria. In the mid-1980s, Syvanen predicted that lateral gene transfer existed, had biological significance, and was involved in shaping evolutionary history from the beginning of life on Earth.
As Jian, Rivera and Lake (1999) put it: "Increasingly, studies of genes and genomes are indicating that considerable horizontal transfer has occurred between prokaryotes" (see also Lake and Rivera, 2007). The phenomenon appears to have had some significance for unicellular eukaryotes as well. As Bapteste et al. (2005) observe, "additional evidence suggests that gene transfer might also be an important evolutionary mechanism in protist evolution."
There is some evidence that even higher plants and animals have been affected and this has raised concerns for safety. Grafting of one plant to another can transfer chloroplasts (organelles in plant cells that conduct photosynthesis), mitochondrial DNA, and the entire cell nucleus containing the genome to potentially make a new species. Some Lepidoptera (e.g. monarch butterflies and silkworms) have been genetically modified by horizontal gene transfer from the wasp bracovirus. Bites from the insect Reduviidae (assassin bug) can, via a parasite, infect humans with the trypanosomal Chagas disease, which can insert its DNA into the human genome. It has been suggested that lateral gene transfer to humans from bacteria may play a role in cancer.
Richardson and Palmer (2007) state: "Horizontal gene transfer (HGT) has played a major role in bacterial evolution and is fairly common in certain unicellular eukaryotes. However, the prevalence and importance of HGT in the evolution of multicellular eukaryotes remain unclear."
Due to the increasing amount of evidence suggesting the importance of these phenomena for evolution (see below) molecular biologists such as Peter Gogarten have described horizontal gene transfer as "A New Paradigm for Biology".
Some have argued that the process may be a hidden hazard of genetic engineering as it could allow transgenic DNA to spread from species to species.
Mechanism
There are several mechanisms for horizontal gene transfer:
A transposon (jumping gene) is a mobile segment of DNA that can sometimes pick up a resistance gene and insert it into a plasmid or chromosome, thereby inducing horizontal gene transfer of antibiotic resistance.
Inference
Horizontal gene transfer is typically inferred using bioinformatic methods, either by identifying atypical sequence signatures ("parametric" methods) or by identifying strong discrepancies between the evolutionary history of particular sequences compared to that of their hosts.
Viruses
The virus called Mimivirus infects amoebae. Another virus, called Sputnik, also infects amoebae, but it cannot reproduce unless mimivirus has already infected the same cell. "Sputnik's genome reveals further insight into its biology. Although 13 of its genes show little similarity to any other known genes, three are closely related to mimivirus and mamavirus genes, perhaps cannibalized by the tiny virus as it packaged up particles sometime in its history. This suggests that the satellite virus could perform horizontal gene transfer between viruses, paralleling the way that bacteriophages ferry genes between bacteria." Horizontal transfer is also seen between geminiviruses and tobacco plants.
Prokaryotes
Horizontal gene transfer is common among bacteria, even among very distantly related ones. This process is thought to be a significant cause of increased drug resistance when one bacterial cell acquires resistance, and the resistance genes are transferred to other species. Transposition and horizontal gene transfer, along with strong natural selective forces have led to multi-drug resistant strains of S. aureus and many other pathogenic bacteria. Horizontal gene transfer also plays a role in the spread of virulence factors, such as exotoxins and exoenzymes, amongst bacteria. A prime example concerning the spread of exotoxins is the adaptive evolution of Shiga toxins in E. coli through horizontal gene transfer via transduction with Shigella species of bacteria. Strategies to combat certain bacterial infections by targeting these specific virulence factors and mobile genetic elements have been proposed. For example, horizontally transferred genetic elements play important roles in the virulence of E. coli, Salmonella, Streptococcus and Clostridium perfringens.
In prokaryotes, restriction-modification systems are known to provide immunity against horizontal gene transfer and in stabilizing mobile genetic elements. Genes encoding restriction modification systems have been reported to move between prokaryotic genomes within mobile genetic elements such as plasmids, prophages, insertion sequences/transposons, integrative conjugative elements (ICEs), and integrons. Still, they are more frequently a chromosomal-encoded barrier to MGEs than an MGE-encoded tool for cell infection.
Bacterial transformation
Natural transformation is a bacterial adaptation for DNA transfer (HGT) that depends on the expression of numerous bacterial genes whose products are responsible for this process. In general, transformation is a complex, energy-requiring developmental process. In order for a bacterium to bind, take up and recombine exogenous DNA into its chromosome, it must become competent, that is, enter a special physiological state. Competence development in Bacillus subtilis requires expression of about 40 genes. The DNA integrated into the host chromosome is usually (but with infrequent exceptions) derived from another bacterium of the same species, and is thus homologous to the resident chromosome. The capacity for natural transformation occurs in at least 67 prokaryotic species. Competence for transformation is typically induced by high cell density and/or nutritional limitation, conditions associated with the stationary phase of bacterial growth. Competence appears to be an adaptation for DNA repair. Transformation in bacteria can be viewed as a primitive sexual process, since it involves interaction of homologous DNA from two individuals to form recombinant DNA that is passed on to succeeding generations. Although transduction is the form of HGT most commonly associated with bacteriophages, certain phages may also be able to promote transformation.
Bacterial conjugation
Conjugation in Mycobacterium smegmatis, like conjugation in E. coli, requires stable and extended contact between a donor and a recipient strain, is DNase resistant, and the transferred DNA is incorporated into the recipient chromosome by homologous recombination. However, unlike E. coli high frequency of recombination conjugation (Hfr), mycobacterial conjugation is a type of HGT that is chromosome rather than plasmid based. Furthermore, in contrast to E. coli (Hfr) conjugation, in M. smegmatis all regions of the chromosome are transferred with comparable efficiencies. Substantial blending of the parental genomes was found as a result of conjugation, and this blending was regarded as reminiscent of that seen in the meiotic products of sexual reproduction.
Archaeal DNA transfer
The archaeon Sulfolobus solfataricus, when UV irradiated, strongly induces the formation of type IV pili which then facilitates cellular aggregation. Exposure to chemical agents that cause DNA damage also induces cellular aggregation. Other physical stressors, such as temperature shift or pH, do not induce aggregation, suggesting that DNA damage is a specific inducer of cellular aggregation.
UV-induced cellular aggregation mediates intercellular chromosomal HGT marker exchange with high frequency, and UV-induced cultures display recombination rates that exceed those of uninduced cultures by as much as three orders of magnitude. S. solfataricus cells aggregate preferentially with other cells of their own species. Frols et al. and Ajon et al. suggested that UV-inducible DNA transfer is likely an important mechanism for providing increased repair of damaged DNA via homologous recombination. This process can be regarded as a simple form of sexual interaction.
Another thermophilic species, Sulfolobus acidocaldarius, is able to undergo HGT. S. acidocaldarius can exchange and recombine chromosomal markers at temperatures up to 84oC. UV exposure induces pili formation and cellular aggregation. Cells with the ability to aggregate have greater survival than mutants lacking pili that are unable to aggregate. The frequency of recombination is increased by DNA damage induced by UV-irradiation and by DNA damaging chemicals.
The ups operon, containing five genes, is highly induced by UV irradiation. The proteins encoded by the ups operon are employed in UV-induced pili assembly and cellular aggregation leading to intercellular DNA exchange and homologous recombination. Since this system increases the fitness of S. acidocaldarius cells after UV exposure, Wolferen et al. considered that transfer of DNA likely takes place in order to repair UV-induced DNA damages by homologous recombination.
Eukaryotes
"Sequence comparisons suggest recent horizontal transfer of many genes among diverse species including across the boundaries of phylogenetic 'domains'. Thus determining the phylogenetic history of a species can not be done conclusively by determining evolutionary trees for single genes."
Horizontal transposon transfer
Horizontal transposon transfer (HTT) refers to the passage of pieces of DNA that are characterized by their ability to move from one locus to another between genomes by means other than parent-to-offspring inheritance. Horizontal gene transfer has long been thought to be crucial to prokaryotic evolution, but there is a growing amount of data showing that HTT is a common and widespread phenomenon in eukaryote evolution as well. On the transposable element (TE) side, spreading between genomes via horizontal transfer may be viewed as a strategy to escape purging due to purifying selection, mutational decay and/or host defense mechanisms.
HTT can occur with any type of transposable elements, but DNA transposons and LTR retroelements are more likely to be capable of HTT because both have a stable, double-stranded DNA intermediate that is thought to be sturdier than the single-stranded RNA intermediate of non-LTR retroelements, which can be highly degradable. Non-autonomous elements may be less likely to transfer horizontally compared to autonomous elements because they do not encode the proteins required for their own mobilization. The structure of these non-autonomous elements generally consists of an intronless gene encoding a transposase protein, and may or may not have a promoter sequence. Those that do not have promoter sequences encoded within the mobile region rely on adjacent host promoters for expression. Horizontal transfer is thought to play an important role in the TE life cycle.
HTT has been shown to occur between species and across continents in both plants and animals (Ivancevic et al. 2013), though some TEs have been shown to more successfully colonize the genomes of certain species over others. Both spatial and taxonomic proximity of species has been proposed to favor HTTs in plants and animals. It is unknown how the density of a population may affect the rate of HTT events within a population, but close proximity due to parasitism and cross contamination due to crowding have been proposed to favor HTT in both plants and animals. Successful transfer of a transposable element requires delivery of DNA from donor to host cell (and to the germ line for multi-cellular organisms), followed by integration into the recipient host genome. Though the actual mechanism for the transportation of TEs from donor cells to host cells is unknown, it is established that naked DNA and RNA can circulate in bodily fluid. Many proposed vectors include arthropods, viruses, freshwater snails (Ivancevic et al. 2013), endosymbiotic bacteria, and intracellular parasitic bacteria. In some cases, even TEs facilitate transport for other TEs.
The arrival of a new TE in a host genome can have detrimental consequences because TE mobility may induce mutation. However, HTT can also be beneficial by introducing new genetic material into a genome and promoting the shuffling of genes and TE domains among hosts, which can be co-opted by the host genome to perform new functions. Moreover, transposition activity increases the TE copy number and generates chromosomal rearrangement hotspots. HTT detection is a difficult task because it is an ongoing phenomenon that is constantly changing in frequency of occurrence and composition of TEs inside host genomes. Furthermore, few species have been analyzed for HTT, making it difficult to establish patterns of HTT events between species. These issues can lead to the underestimation or overestimation of HTT events between ancestral and current eukaryotic species.
Artificial horizontal gene transfer
Genetic engineering is essentially horizontal gene transfer, albeit with synthetic expression cassettes. The Sleeping Beauty transposon system (SB) was developed as a synthetic gene transfer agent that was based on the known abilities of Tc1/mariner transposons to invade genomes of extremely diverse species. The SB system has been used to introduce genetic sequences into a wide variety of animal genomes. (See also Gene therapy.)
Importance in evolution
Horizontal gene transfer is a potential confounding factor in inferring phylogenetic trees based on the sequence of one gene. For example, given two distantly related bacteria that have exchanged a gene a phylogenetic tree including those species will show them to be closely related because that gene is the same even though most other genes are dissimilar. For this reason it is often ideal to use other information to infer robust phylogenies such as the presence or absence of genes or, more commonly, to include as wide a range of genes for phylogenetic analysis as possible.
For example, the most common gene to be used for constructing phylogenetic relationships in prokaryotes is the 16S ribosomal RNA gene since its sequences tend to be conserved among members with close phylogenetic distances, but variable enough that differences can be measured. However, in recent years it has also been argued that 16s rRNA genes can also be horizontally transferred. Although this may be infrequent, the validity of 16s rRNA-constructed phylogenetic trees must be reevaluated.
Biologist Johann Peter Gogarten suggests "the original metaphor of a tree no longer fits the data from recent genome research" therefore "biologists should use the metaphor of a mosaic to describe the different histories combined in individual genomes and use the metaphor of a net to visualize the rich exchange and cooperative effects of HGT among microbes". There exist several methods to infer such phylogenetic networks.
Using single genes as phylogenetic markers, it is difficult to trace organismal phylogeny in the presence of horizontal gene transfer. Combining the simple coalescence model of cladogenesis with rare HGT horizontal gene transfer events suggest there was no single most recent common ancestor that contained all of the genes ancestral to those shared among the three domains of life. Each contemporary molecule has its own history and traces back to an individual molecule cenancestor. However, these molecular ancestors were likely to be present in different organisms at different times."
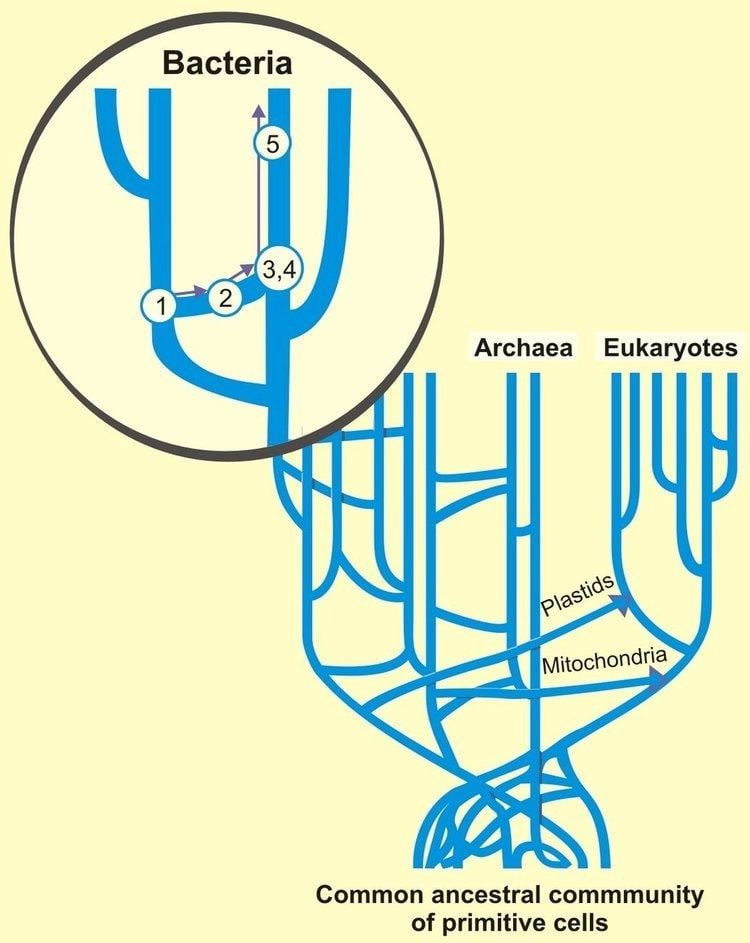
Challenge to the tree of life
Horizontal gene transfer poses a possible challenge to the concept of the last universal common ancestor (LUCA) at the root of the tree of life first formulated by Carl Woese, which led him to propose the Archaea as a third domain of life. Indeed, it was while examining the new three-domain view of life that horizontal gene transfer arose as a complicating issue: Archaeoglobus fulgidus was seen as an anomaly with respect to a phylogenetic tree based upon the encoding for the enzyme HMGCoA reductase—the organism in question is a definite Archaean, with all the cell lipids and transcription machinery that are expected of an Archaean, but whose HMGCoA genes are of bacterial origin. Scientists are broadly agreed on symbiogenesis, that mitochondria in eukaryotes derived from alpha-proteobacterial cells and that chloroplasts came from ingested cyanobacteria, and other gene transfers may have affected early eukaryotes. (In contrast, multicellular eukaryotes have mechanisms to prevent horizontal gene transfer, including separated germ cells.) If there had been continued and extensive gene transfer, there would be a complex network with many ancestors, instead of a tree of life with sharply delineated lineages leading back to a LUCA. However, a LUCA can be identified, so horizontal transfers must have been relatively limited.
Genes
There is evidence for historical horizontal transfer of the following genes:
