Scientific name Alphaproteobacteria | ||
 | ||
Lower classifications | ||
Medical vocabulary what does alphaproteobacteria mean
Alphaproteobacteria is a class of Bacteria in the phylum Proteobacteria (See also bacterial taxonomy). Its members are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all Proteobacteria, its members are Gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable.
Contents
- Medical vocabulary what does alphaproteobacteria mean
- Characteristics
- Evolution and genomics
- Phylogeny
- Natural genetic transformation
- References
Characteristics
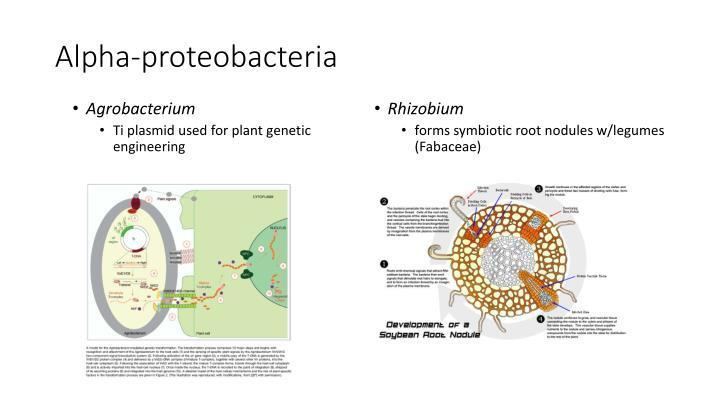
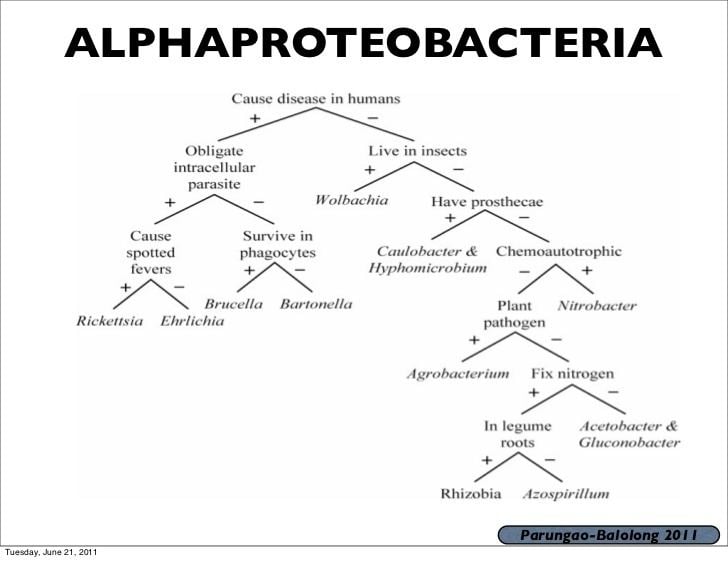
The Alphaproteobacteria is a diverse taxon and comprises several phototrophic genera, several genera metabolising C1-compounds (e.g., Methylobacterium spp.), symbionts of plants (e.g., Rhizobium spp.), endosymbionts of arthropods (Wolbachia) and intracellular pathogens (e.g. Rickettsia). Moreover, the class includes (as an extinct member) the protomitochondrion, the bacterium that was engulfed by the eukaryotic ancestor and gave rise to the mitochondria, which are organelles in eukaryotic cells (See endosymbiotic theory). A species of technological interest is Rhizobium radiobacter (formerly Agrobacterium tumefaciens): scientists often use this species to transfer foreign DNA into plant genomes. Aerobic anoxygenic phototrophic bacteria, such as Pelagibacter ubique, are alphaproteobacteria that are a widely distributed marine plankton that may constitute over 10% of the open ocean microbial community.
Evolution and genomics
There is some disagreement on the phylogeny of the orders, especially for the location of the Pelagibacterales, but overall there is some consensus. This issue stems form the large difference in gene content (e.g. genome streamlining in Pelagibacter ubique) and the large difference in GC-richness between members of several order. Specifically,Pelagibacterales, Rickettsiales and Holosporales contains species with AT-rich genomes. It has been argued that it could be a case of convergent evolution that would result in an artefactual clustering. However, several studies disagree,. Furthermore, it has been found that the GC-content of ribosomal RNA, the traditional phylogenetic marker, little reflects the GC-content of the genome: for example, members of the Holosporales have a much higher ribosomal GC-content than members of the Pelagibacterales and Rickettsiales, which have similarly low genomic GC-content, because they are more closely related to species with high genomic GC-contents than to members of the latter two orders
The Class Alphaproteobacteria is divided into three subclasses Magnetococcidae, Rickettsidae and Caulobacteridae. The basal group is Magnetococcidae, which is composed by a large diversity of magnetotactic bacteria, but only one is described, Magnetococcus marinus. The Rickettsidae is composed of the intracellular Rickettsiales and the free-living Pelagibacterales. The Caulobacteridae is composed of the Holosporales, Rhodospirillales, Sphingomonadales, Rhodobacterales, Caulobacterales, Kiloniellales, Kordiimonadales, Parvularculales and Sneathiellales.

Comparative analyses of the sequenced genomes have also led to discovery of many conserved indels in widely distributed proteins and whole proteins (i.e. signature proteins) that are distinctive characteristics of either all Alphaproteobacteria, or their different main orders (viz. Rhizobiales, Rhodobacterales, Rhodospirillales, Rickettsiales, Sphingomonadales and Caulobacterales) and families (viz. Rickettsiaceae, Anaplasmataceae, Rhodospirillaceae, Acetobacteraceae, Bradyrhiozobiaceae, Brucellaceae and Bartonellaceae). These molecular signatures provide novel means for the circumscription of these taxonomic groups and for identification/assignment of new species into these groups. Phylogenetic analyses and conserved indels in large numbers of other proteins provide evidence that Alphaproteobacteria have branched off later than most other phyla and Classes of Bacteria except Betaproteobacteria and Gammaproteobacteria.
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) and the phylogeny is based on 16S rRNA-based LTP release 106 by 'The All-Species Living Tree' Project
Notes:
♠ Strains found at the National Center for Biotechnology Information (NCBI) but not listed in the List of Prokaryotic names with Standing in Nomenclature (LSPN)
Natural genetic transformation
Although only a few studies have been reported on natural genetic transformation in the Alphaproteobacteria, this process has been described in Agrobacterium tumefaciens, Methylobacterium organophilum, and Bradyrhizobium japonicum. Natural genetic transformation is a sexual process involving DNA transfer from one bacterial cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination.
