 | ||
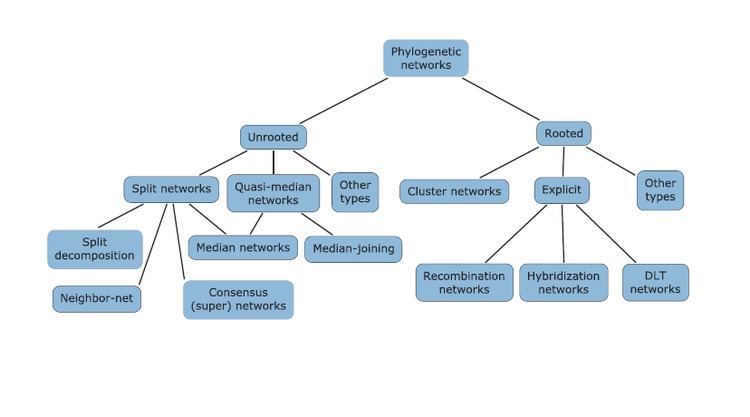
A phylogenetic network or reticulation is any graph used to visualize evolutionary relationships (either abstractly or explicitly) between nucleotide sequences, genes, chromosomes, genomes, or species. They are employed when reticulation events such as hybridization, horizontal gene transfer, recombination, or gene duplication and loss are believed to be involved. They differ from phylogenetic trees by the explicit modeling of richly-linked networks, by means of the addition of hybrid nodes (nodes with two parents) instead of only tree nodes (a hierarchy of nodes, each with only one parent). Phylogenetic trees are a subset of phylogenetic networks. Phylogenetic networks can be inferred and visualised with software such as SplitsTree and, more recently, Dendroscope. A standard format for representing phylogenetic networks is a variant of Newick format which is extended to support networks as well as trees.
Contents
Many kinds and subclasses of phylogenetic networks have been defined based on the biological phenomenon they represent or which data they are built from (hybridization networks, usually built from rooted trees, recombination networks from binary sequences, median networks from a set of splits, optimal realizations and reticulograms from a distance matrix), or restrictions to get computationally tractable problems (galled trees, and their generalizations level-k phylogenetic networks, tree-child or tree-sibling phylogenetic networks).
Microevolution
Phylogenetic trees also have trouble depicting microevolutionary events, for example the geographical distribution of muskrat or fish populations of a given species among river networks, because there is no species boundary to prevent gene flow between populations. Therefore, a more general phylogenetic network better depicts these situations.
