Entrez 8450 | Ensembl ENSG00000158290 | |
 | ||
Aliases CUL4B, CUL-4B, MRXHF2, MRXS15, MRXSC, SFM2, cullin 4B External IDs MGI: 1919834 HomoloGene: 2660 GeneCards: CUL4B | ||
Gene music using protein sequence of cul4b cullin 4b
Cullin-4B is a protein that in humans is encoded by the CUL4B gene which is located on the X chromosome. CUL4B has high sequence similarity with CUL4A, with which it shares certain E3 ubiquitin ligase functions. CUL4B is largely expressed in the nucleus and regulates several key functions including: cell cycle progression, chromatin remodeling and neurological and placental development in mice. In humans, CUL4B has been implicated in X-linked intellectual disability and is frequently mutated in pancreatic adenocarcinomas and a small percentage of various lung cancers. Viruses such as HIV can also co-opt CUL4B-based complexes to promote viral pathogenesis. CUL4B complexes containing Cereblon are also targeted by the teratogenic drug thalidomide.
Contents
- Gene music using protein sequence of cul4b cullin 4b
- Structure
- Cell cycle regulation and chromatin remodeling
- Mammalian embryonic development
- Neurological development
- X linked intellectual disability
- Viral pathogenesis
- Cancer
- Thalidomide treatment
- Interactions and substrates
- References
Structure
Human CUL4B is 913 amino acids long and shares a high degree of sequence identity (84%) with CUL4A with the exception of its unique N-terminal region. The extreme N-terminus of CUL4B is disordered and, currently, it is unclear what structural and functional qualities it possesses. CUL4B binds to the beta-propeller of the DDB1 adaptor protein which interacts with numerous DDB1-CUL4-Associated Factors (DCAFs). This interaction is crucial for the recruitment of substrates to the ubiquitin ligase complex. At the C-terminal end, CUL4B interacts with the RBX1/ROC1 protein via its RING domain. RBX1 is a core component of Cullin-RING ubiquitin ligase (CRL) complexes and functions to recruit E2 ubiquitin conjugating enzymes. Therefore, the C-terminus of CUL4B - along with RBX1 and activated E2 enzymes - compose the catalytic core of CRL4B complexes. CUL4B is also modified by covalent attachment of a NEDD8 molecule at a highly conserved lysine residue in the C-terminal region. This modification appears to induce conformational changes which promotes flexibility in the RING domain of cullin proteins and enhanced ubiquitin ligase activity.
Cell cycle regulation and chromatin remodeling
CUL4B-based E3 ubiquitin ligase complexes often demonstrate overlapping activity with CUL4A-based complexes. Both CRL4 complexes utilize Cdt2 and the DNA processivity factor PCNA to induce the ubiquitination and degradation of replication licensing factor Cdt1 and cyclin-dependent kinase inhibitor p21 in a proteasome-dependent manner. CRL4Cdt2 also degrades PCNA-bound PR-Set7/SET8, which is a histone 4 methyltransferase, and the p12 subunit of DNA polymerase δ, which is crucial for DNA replication. As a result, CRL4 complexes are able to control the onset of DNA replication, chromatin remodeling and progression through the cell cycle.
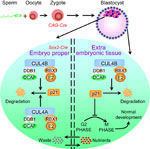
Mammalian embryonic development
Loss of Cul4b in mice causes embryonic lethality and defects in placental development. The extra-embryonic tissue of these developing mice also showed increased rates of apoptosis and a decrease in cell proliferation. When Cul4b deletion was limited to the epiblast (only in Sox2-expressing tissue), it was possible to generate living mice.
Neurological development
Mice that do not express CUL4B in epiblast tissue demonstrate normal brain morphology but decrease number of parvalbumin (PV)-positive GABAergic interneurons - particularly in the dentate gyrus. In these mice, certain dendritic features of hippocampal neurons were also affected by Cul4b loss, which may explain the observed increases in epilectic susceptibility and spatial learning defects. These phenotypes resembled features seen in patients with X-linked intellectual disability (see below).
X-linked intellectual disability
Loss-of-function CUL4B mutation events have been discovered in numerous patients with X-linked intellectual disability , which is characterized by aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus and tremor. CUL4B mutations have also been associated with malformations of cortical development.
Viral pathogenesis
After HIV infects a cell, the virus "hijacks" either the CUL4B-DDB1 complex or the CUL4A-DDB1 complex via the same mechanism. Essentially, HIV proteins such as Vpr and Vpx bind to VPRBP (a DDB1-binding substrate receptor protein) and induce the ubiquitination and degradation of SAMHD1 and UNG2 to promote viral replication. These proteins are not degraded by CRL4 complexes in the absence of virus.
Cancer
According to data from The Cancer Genome Atlas, CUL4B is mutated in 21% of pancreatic carcinomas with a recurring truncating mutation at amino acid 143. CUL4B is also mutated or amplified in 3-5% of lung cancers. The significance of these observed mutations has not been determined.
Thalidomide treatment
In 2010, Ito et al. reported that Cereblon, a DCAF protein, was a major target of the teratogenic compound thalidomide. Thalidomide and other derivatives such as pomalidomide and lenalidomide are known as immunomodulatory drugs (or IMiDs) and have been investigated as therapeutic agents for autoimmune diseases and several cancers - particularly myelomas. Recent reports show that IMiDs bind to CRL4CRBN and promote the degradation of IKZN1 and IKZN3 transcription factors, which are not normally targeted by CRL4 complexes.
Interactions and substrates
Human CUL4B forms direct interactions with:
Human CUL4B-DDB1-RBX1 complexes promote the ubiquitination of:
†protein is a CRL4 substrate only when directed by viral proteins
§protein is a CRL4 substrate only when directed by IMiDs
