Aliases CUL4A Ensembl ENSG00000139842 | Entrez 8451 | |
 | ||
External IDs MGI: 1914487 HomoloGene: 81724 GeneCards: CUL4A | ||
Gene music using protein sequence of cul4a cullin 4a
Cullin-4A is a protein that in humans is encoded by the CUL4A gene. CUL4A belongs to the cullin family of ubiquitin ligase proteins and is highly homologous to the CUL4B protein. CUL4A regulates numerous key processes such as DNA repair, chromatin remodeling, spermatogenesis, haematopoiesis and the mitotic cell cycle. As a result, CUL4A has been implicated in several cancers and the pathogenesis of certain viruses including HIV. Interestingly, a component of a CUL4A complex, Cereblon, was discovered to be a major target of the teratogenic agent thalidomide.
Contents
- Gene music using protein sequence of cul4a cullin 4a
- Structure
- DNA damage and repair
- Chromatin remodeling
- Regulation of the cell cycle and DNA replication
- Haematopoiesis
- Spermatogenesis and meiosis
- Cancer
- Viral pathogenesis
- Thalidomide treatment
- Interactions and substrates
- References
Structure
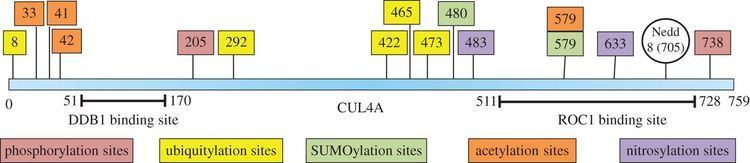
CUL4A protein is 759 amino acids long and forms an extended, rigid structure primarily consisting of alpha-helices. At the N-terminus, CUL4A binds to the beta-propeller of the DDB1 adaptor protein which interacts with numerous DDB1-CUL4-Associated Factors (DCAFs). As a result, the N-terminus is crucial for the recruitment of substrates for the ubiquitin ligase complex. At the C-terminal end, CUL4A interacts with the RBX1/ROC1 protein via its RING domain. RBX1 is a core component of Cullin-RING ubiquitin ligase (CRL) complexes and functions to recruit E2 ubiquitin conjugating enzymes. Therefore, the C-terminus of CUL4A - along with RBX1 and activated E2 enzymes - compose the catalytic core of CRL4 complexes. CUL4A is also modified by covalent attachment of a NEDD8 molecule at a highly conserved lysine residue in the C-terminal region. This modification appears to induce conformational changes which promotes flexibility in the RING domain of cullin proteins and enhanced ubiquitin ligase activity.
Overall, CRL4A complexes have a modular structure which allows for sophisticated regulation by the cell and influence over numerous substrates and processes in the cell. Although the individual parts vary, all cullin-based ubiquitin ligases exhibit these characteristics.
DNA damage and repair
The DDB1 adaptor protein was initially characterized as the large subunit of a heterodimeric complex (UV-DDB) that was found to recognize damaged DNA and participate in a form of repair known as nucleotide excision repair (NER). The smaller subunit of this Damaged DNA Binding protein complex is known as DDB2 and is able to directly bind DNA lesions associated with UV-irradiation. DDB2 is a DCAF protein and is both a ubiquitination substrate of the CRL4 complex and also serves as an E3 ligase protein for other substrates such as XPC and histones (see next section) near the damage site. Due to its ubiquitination of DNA damage-recognizing proteins DDB2 and XPC, CUL4A has been described as a negative regulator of NER activity. In addition to the "global" type of NER, the CRL4A complex also appears to play a role in "transcription-coupled" NER in conjunction with the Cockayne Syndrome A protein. CRL4A complexes appear to be activated by certain types of DNA damage (most notably, UV-irradiation) and several substrates are preferentially ubiquitinated after DNA damage induction.
Chromatin remodeling
CUL4A's role in modifying chromatin is largely related to DNA repair activities and occurs after DNA damage induction. Both CUL4A and its closely related homolog CUL4B may ubiquitinate histones H2A, H3 and H4. The yeast homolog of CUL4A, Rtt101, ubiquitinates histone H3 and promotes nucleosome assembly and CRL4A complexes perform similar functions in human cells. CRL4 complexes also affect histone methylation events and chromatin structure through regulation of histone methyltransferases. The histone H4 monomethylase PR-Set7/SET8 is ubiquitinated on chromatin by CRL4(Cdt2) complexes during S phase and following DNA damage in a PCNA-dependent manner.
Regulation of the cell cycle and DNA replication
CRL4A complexes regulate entry into the DNA synthesis phase, or S phase, of the mitotic cycle by regulating protein expression levels of the replication licencing factor protein Cdt1 and cyclin-dependent kinase inhibitor p21. In both cases, CRL4A utilizes Cdt2 as the DCAF to bind both substrates in a PCNA-dependent manner. During unperturbed cell cycle progression, ubiquitination and downregulation of these proteins by CRL4ACdt2 occurs at the onset of DNA replication. DNA damage such as UV irradiation also induces CRL4ACdt2-mediated destruction of those proteins. Interestingly, both substrates are also regulated by the SCFSkp2 complex.
CRL4-mediated destruction of p21 relieves cyclin E-Cdk2 inhibition and promotes S phase entry. Loss of Cdt2 expression increases p21 expression in cells and stabilizes p21 following UV-irradiation. CUL4A deletion results in delayed S phase entry in mouse embryonic fibroblasts, which is rescued by deletion of p21.
After promoting initiation of eukaryotic DNA replication at the origin, Cdt1 is inactivated by Geminin and targeted for degradation by the SCFSkp2 and CRL4Cdt2 complexes. Cdt1 expression is stabilized by RNAi-mediated knockdown of DDB1 or both CUL4A and CUL4B, which suggests redundant or overlapping function of the two CUL4 proteins for Cdt1 regulation. Only reduction of Geminin expression seems to induce re-replication in Cdt1-overexpressing cells.
CRL4s also utilize Cdt2 and PCNA to degrade the p12 subunit of DNA polymerase δ during S phase and after UV irradiation.
Haematopoiesis
CRL4A complexes appear to induce the degradation of numerous members of the HOX transcription family, which are essential regulators of haematopoiesis. The first member of the HOX family identified as a target of CRL4A-mediated degradation is HOXA9, which is essential for haematopoietic stem cell maintenance and has been implicated in a subset of myeloid leukemias. The HOXA9 degron lies within the homeodomain, which is crucial for DNA binding. Sequence alignment studies showed that there is a highly conserved "LEXE" motif within helix one of the homeodomain. When multiple amino acids within this motif were mutated, HOXB4 became resistant to CRL4A-mediated degradation. The substrate receptor, or DCAF, required for HOX protein degradation remains unknown.
Spermatogenesis and meiosis
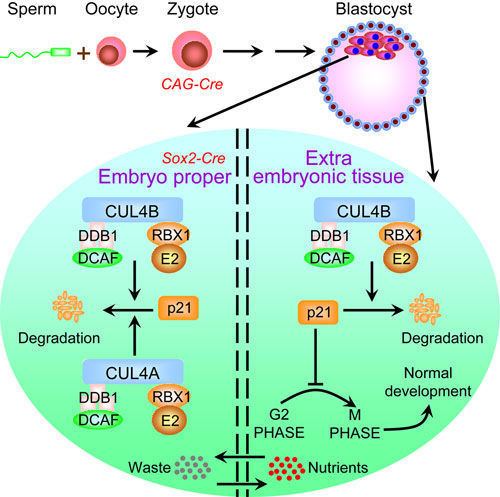
The Cul4a gene is required for normal spermatogenesis and meiosis in male germ cells of mice. Cul4a−/− males produce abnormal sperm and are infertile. While both CUL4A and CUL4B are expressed in male gametes, CUL4A is highly expressed in pachytenes and diplotenes. It is at these stages that CUL4A-deficient male germ cells exhibit high levels of apoptosis, improper DNA repair and accumulation of the CRL4 substrate Cdt1.
Cancer
The chromosomal region ch13q34 which contains the CUL4A gene is amplified in 3-6% of certain carcinomas including: breast, uterine, lung, stomach and colorectal cancers. CUL4A is also mutated or amplified in about 4% of melanomas (although the mutations are dispersed and individual mutations occur sporadically).
In mouse models, Cul4a knockout resulted in pronounced resistance to UV-induced skin carcinogenesis. Cre-induced Cul4a overexpression in mouse lung tissue promoted hyperplasia.
Due to the observed amplification of CUL4A in several carcinomas and the fact that CRL4 complexes target multiple DNA repair and tumor suppressor genes, CUL4A can be considered an oncogene in certain contexts.
Viral pathogenesis
Due to its robust expression (particularly during DNA replication) and modular nature, CRL4A complexes can be co-opted or "hijacked" to promote viral proliferation in mammalian cells.
Certain paramyxoviruses avoid the interferon response in cells by targeting STAT1 and disrupting signaling. Simian virus 5 and type II human parainfluenza virus express a protein, named "V", which acts as a substrate receptor and bridges an interaction between DDB1 and STAT proteins (the structure of the CRL4ASV5V complex is pictured in the inset) - thus inducing STAT1 ubiquitination and degradation
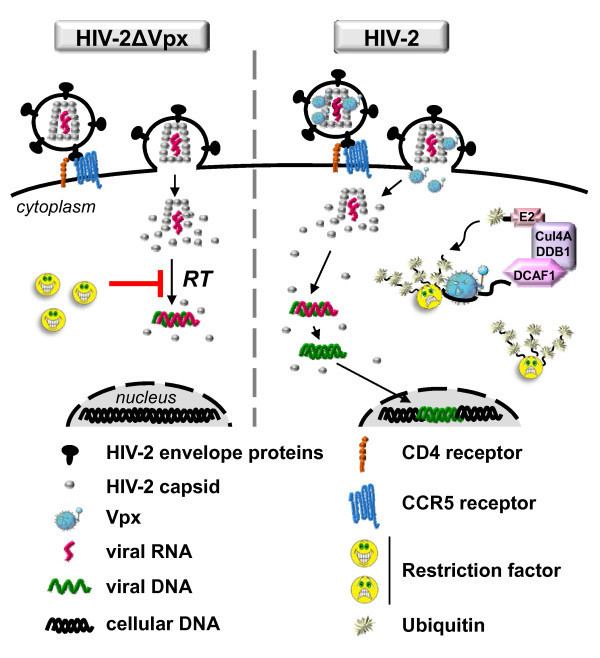
DCAF1 is also named VPRBP due to its interaction with HIV-1 protein Vpr. Although DCAF1/VPRBP appears to have a crucial function in tumor suppression, DNA replication and embryonic development, HIV-1 "hijacks" the ubiquitin ligase complex to induce arrest of the cell cycle in G2 phase. The CRL4ADCAF1-Vpr induces ubiquitination of the nuclear isoform of uracil-DNA glycosylase. HIV-2 also appears to utilize CRL4ADCAF1 via Vpx protein-induced destruction of a lentivirus-inhibiting deoxynucleoside triphosphohydrolase named SAMHD1.
Thalidomide treatment
In 2010, Ito et al. reported that Cereblon, a DCAF protein, was a major target of the teratogenic compound thalidomide. Thalidomide and other derivatives such as pomalidomide and lenalidomide are known as immunomodulatory drugs (or IMiDs) and have been investigated as therapeutic agents for autoimmune diseases and several cancers - particularly myelomas. Recent reports show that IMiDs bind to CRL4CRBN and promote the degradation of IKZN1 and IKZN3 transcription factors, which are not normally targeted by CRL4 complexes.
Interactions and substrates
Human CUL4A forms direct interactions with:
Human CUL4A-DDB1-RBX1 complexes promote the ubiquitination of:
†protein is a CRL4A substrate only when directed by viral proteins
§protein is a CRL4A substrate only when directed by IMiDs
