Group Group II (ssDNA) Genus Microvirus Rank Species | Family Microviridae Higher classification Microvirus | |
 | ||
Scientific name Enterobacteria phage phiX174 Similar Bacteriophage MS2, Enterobacteria phage T4, M13 bacteriophage, Microviridae, Lambda phage | ||
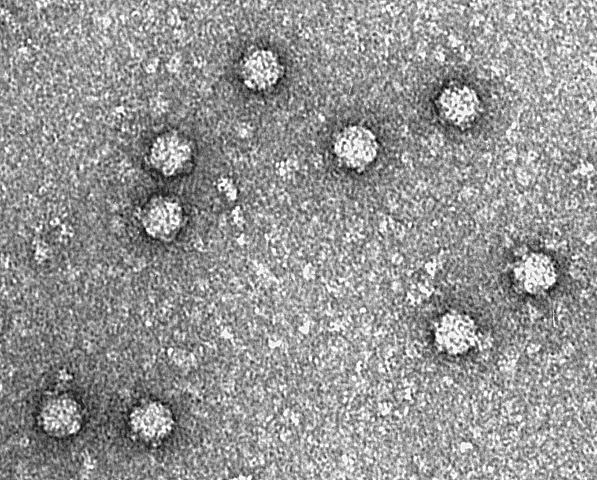
The phi X 174 (or ΦX174) bacteriophage is a virus and was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA. Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. Recently, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.
Virology
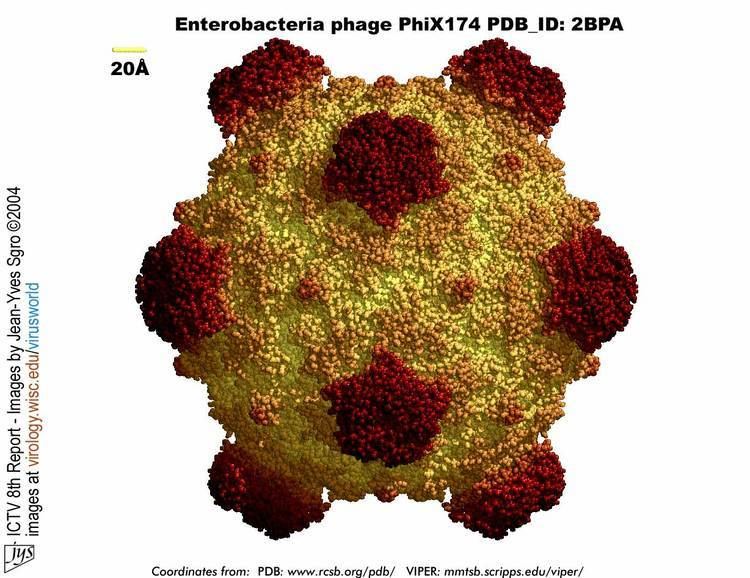
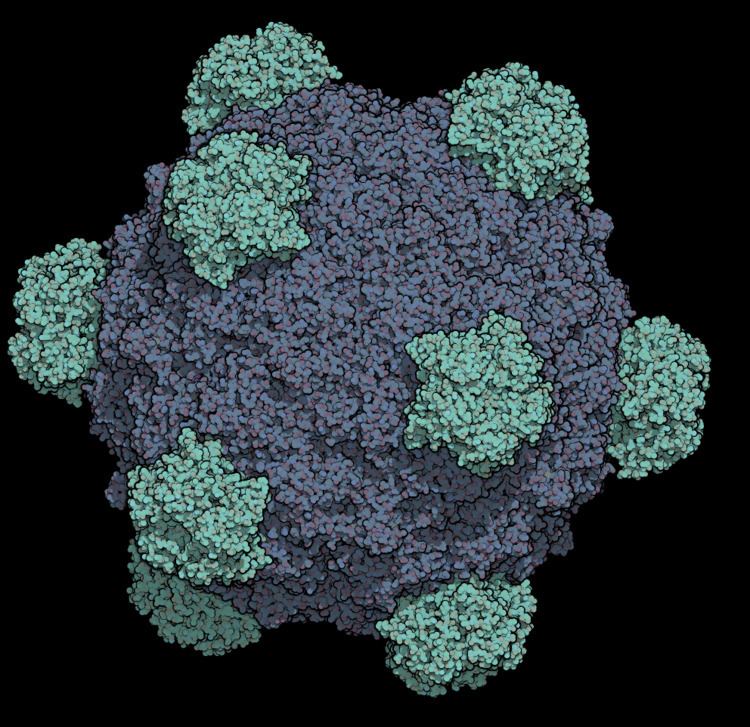
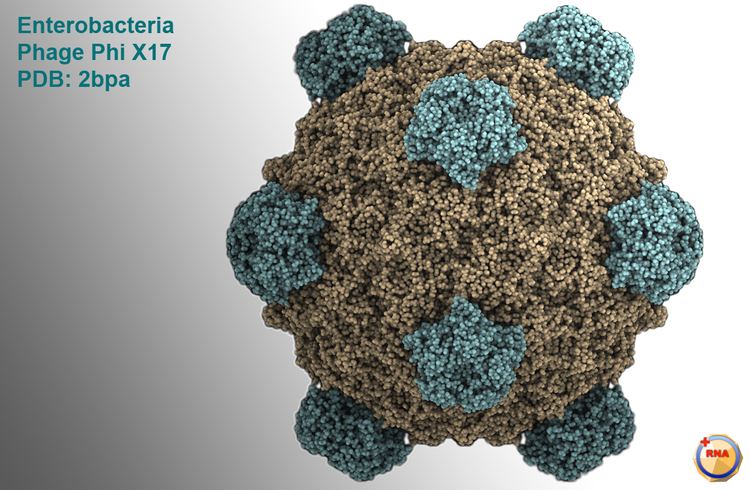
This bacteriophage has a [+] circular single-stranded DNA genome of 5386 nucleotides encoding 11 proteins. Of these 11 genes, only 8 are essential to viral morphogenesis. The GC-content is 44% and 95% of nucleotides belong to coding genes.
Infection begins when G protein binds to lipopolysaccharides on the bacterial host cell surface. H protein (or the DNA Pilot Protein) pilots the viral genome through the bacterial membrane of E.coli bacteria (Jazwinski et al. 1975) most likely via a predicted N-terminal transmembrane domain helix (Tusnady and Simon, 2001). However, it has become apparent that H protein is a multifunctional protein (Cherwa, Young and Fane, 2011). This is the only viral capsid protein of ΦX174 to lack a crystal structure for a couple of reasons. It has low aromatic content and high glycine content, making the protein structure very flexible and in addition, individual hydrogen atoms (the R group for glycines) are difficult to detect in protein crystallography. Additionally, H protein induces lysis of the bacterial host at high concentrations as the predicted N-terminal transmembrane helix easily pokes holes through the bacterial wall. By bioinformatics, this protein contains four predicted coiled-coil domains which has a significant homology to known transcription factors. Additionally, it was determined by Ruboyianes et al. (2009) that de novo H protein was required for optimal synthesis of other viral proteins. Interestingly, mutations in H protein that prevent viral incorporation, can be overcome when excess amounts of Protein B, the internal scaffolding protein, are supplied.
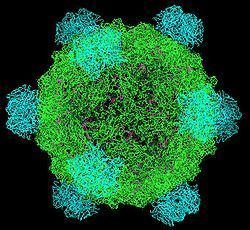
The DNA is ejected through a hydrophilic channel at the 5-fold vertex (McKenna et al. 1992). It is understood that H protein resides in this area but experimental evidence has not verified its exact location. Once inside the host bacterium, replication of the [+] ssDNA genome proceeds via negative sense DNA intermediate. This is done as the phage genome supercoils and the secondary structure formed by such supercoiling attracts a primosome protein complex. This translocates once around the genome and synthesizes a [-]ssDNA from the positive original genome. [+]ssDNA genomes to package into viruses are created from this by a rolling circle mechanism. This is the mechanism by which the double stranded supercoiled genome is nicked on the negative strand by a virus-encoded A protein, also attracting a bacterial DNA Polymerase to the site of cleavage. DNAP will use the negative strand as a template to make positive sense DNA. As it translocates around the genome it displaces the outer strand of already-synthesised DNA, which is immediately coated by ssBP proteins. The A protein will cleave the complete genome every time it recognises the origin sequence.
As D protein is the most abundant gene transcript, it is the most protein in the viral procapsid. Similarly, gene transcripts for F, J, and G are more abundant than for H as the stoichiometry for these structural proteins is 5:5:5:1.The primosome are protein complexes which attach/bind the enzyme helicase on the template. primosomes gives RNA primers for DNA synthesis to strands.
