 | ||
An overlapping gene is a gene whose expressible nucleotide sequence partially overlaps with the expressible nucleotide sequence of another gene. In this way, a nucleotide sequence may make a contribution to the function of one or more gene products. Overprinting refers to a type of overlap in which all or part of the sequence of one gene is read in an alternate reading frame from another gene at the same locus. Overprinting has been hypothesized as a mechanism for de novo emergence of new genes from existing sequences, either older genes or previously non-coding regions of the genome. Overprinted genes are particularly common features of the genomic organization of viruses, likely to greatly increase the number of potential expressible genes from a small set of viral genetic information.
Contents
Classification
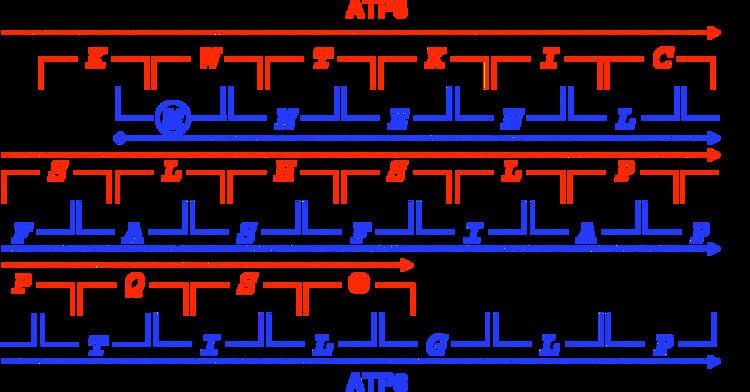
Genes may overlap in a variety of ways and can be classified by their positions relative to each other.
Overlapping genes can also be classified by phases, which describe their relative reading frames:
Evolution
Overlapping genes are particularly common in rapidly evolving genomes, such as those of viruses, bacteria, and mitochondria. They may originate in three ways:
- By extension of an existing open reading frame (ORF) downstream into a contiguous gene due to the loss of a stop codon;
- By extension of an existing ORF upstream into a contiguous gene due to loss of an initiation codon;
- By generation of a novel ORF within an existing one due to a point mutation.
The use of the same nucleotide sequence to encode multiple genes may provide evolutionary advantage due to reduction in genome size and due to the opportunity for transcriptional and translational co-regulation of the overlapping genes. Gene overlaps introduce novel evolutionary constraints on the sequences of the overlap regions.
Origins of new genes
Certain types of gene overlap provide evidence that one of the genes in the pair may have originated de novo, rather than from gene duplication or other mechanisms by which new genes are created. In 1977, Pierre-Paul Grassé proposed the possibility that genes could be generated de novo by mutations in existing genetic sequences to introduce novel ORFs in alternate reading frames; he described the mechanism as overprinting. It was later substantiated by Susumu Ohno, who identified a candidate gene that may have arisen by this mechanism. The mechanism is equally applicable both to generation of a novel ORF overlapping an existing one, and to generation of a novel ORF from previously non-coding DNA. It has since been proposed as a common source of de novo genes, which may make up a significant fraction of so-called "ORFan" or taxonomically restricted genes.
Overprinting is known to be a common source of new genes in virus genomes. Overprinting can be detected bioinformatically by comparison of codon usage statistics between the original and putative overprinted gene; in most cases the overprinted gene is more phylogenetically restricted and is less likely to match the codon usage distribution found in the genome overall. Overprinted genes that exhibit these properties more strongly tend to be evolutionarily more recent.
Taxonomic distribution
Overlapping genes occur in all domains of life, though with varying frequencies. They are especially common in viral genomes.
Viruses
The existence of overlapping genes was first identified in viruses; the first DNA genome ever sequenced, of the bacteriophage ΦX174, contained several examples. Overlapping genes are particularly common in viral genomes. Some studies attribute this observation to selective pressure toward small genome sizes mediated by the physical constraints of packaging the genome in a viral capsid, particularly one of icosahedral geometry. However, other studies dispute this conclusion and argue that the distribution of overlaps in viral genomes is more likely to reflect overprinting as the evolutionary origin of overlapping viral genes. Overprinting is a common source of de novo genes in viruses.
Studies of overprinted viral genes suggest that their protein products tend to be accessory proteins which are not essential to viral proliferation, but contribute to pathogenicity. Overprinted proteins often have unusual amino acid distributions and high levels of intrinsic disorder. In some cases overprinted proteins do have well-defined, but novel, three-dimensional structures; one example is the RNA silencing suppressor p19 found in Tombusviruses, which has both a novel protein fold and a novel binding mode in recognizing siRNAs.
Prokaryotes
Estimates of gene overlap in bacterial genomes typically find that around one third of bacterial genes are overlapped, though usually only by a few base pairs. Most studies of overlap in bacterial genomes find evidence that overlap serves a function in gene regulation, permitting the overlapped genes to be transcriptionally and translationally co-regulated. In prokaryotic genomes, unidirectional overlaps are most common, possibly due to the tendency of adjacent prokaryotic genes to share orientation. Among unidirectional overlaps, long overlaps are more commonly read with a one-nucleotide offset in reading frame (i.e., phase 1) and short overlaps are more commonly read in phase 2. Long overlaps of greater than 60 base pairs are more common for convergent genes; however, putative long overlaps have very high rates of misannotation. Robustly validated examples of long overlaps in bacterial genomes are rare; in the well-studied model organism Escherichia coli, only four gene pairs are well validated as having long, overprinted overlaps.
Eukaryotes
Compared to prokaryotic genomes, eukaryotic genomes are often poorly annotated and thus identifying genuine overlaps is relatively challenging. However, examples of validated gene overlaps have been documented in a variety of eukaryotic organisms, including mammals such as mice and humans. Eukaryotes differ from prokaryotes in distribution of overlap types: while unidirectional (i.e., same-strand) overlaps are most common in prokaryotes, opposite or antiparallel-strand overlaps are more common in eukaryotes. Among the opposite-strand overlaps, convergent orientation is most common. Most studies of eukaryotic gene overlap have found that overlapping genes are extensively subject to genomic reorganization even in closely related species, and thus the presence of an overlap is not always well-conserved. Overlap with older or less taxonomically restricted genes is also a common feature of genes likely to have originated de novo in a given eukaryotic lineage.
