Scientific name Mononegavirales Rank Order | Higher classification Viral Group V | |
 | ||
Lower classifications | ||
The order Mononegavirales is the taxonomic home of numerous related viruses. Members of the order that are commonly known are, for instance, Ebola virus, human respiratory syncytial virus, measles virus, mumps virus, Nipah virus, and rabies virus. All of these viruses cause significant disease in humans. Many very important pathogens of nonhuman animals and plants are also members of this order.
Contents
Use of term
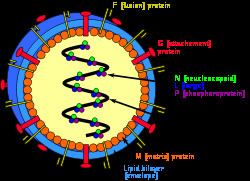
The order Mononegavirales is a virological taxon that was created in 1991 and emended in 1995, 1997, 2000, 2005, 2011, and 2016. The name Mononegavirales is derived from the Greek adjective μóνος [monos] (alluding to the monopartite and single-stranded genomes of most mononegaviruses), the Latin verb negare (alluding to the negative polarity of these genomes), and the taxonomic suffix -virales (denoting a viral order). The order currently includes the eight virus families Bornaviridae, Mymonaviridae, Filoviridae, Nyamiviridae, Paramyxoviridae, Pneumoviridae, Rhabdoviridae, and Sunviridae.
Note
Mononegavirales is pronounced ˌmɒnəˌnɛgəviː’rɑ:lɨz (IPA) or mo-nuh-ne-guh-vee-rah-liz in English phonetic notation. According to the rules for taxon naming established by the International Committee on Taxonomy of Viruses (ICTV), the name Mononegavirales is always to be capitalized, italicized, and never abbreviated. The names of the order's physical members (mononegaviruses/mononegavirads) are to be written in lower case, are not italicized, and used without articles.
Order inclusion criteria
A virus is a member of the order Mononegavirales if
Order organization
The order includes eight accepted families that include numerous genera, consisting of many different species. The order has expanded considerably during recent years due to the discovery of many novel agents that were found to be phylogenetically diverse from already known mononegaviruses. Novel taxa (genera and/or species) had to be proposed, some of which have by now been accepted by the ICTV, and others that are in various stages of the suggestion/proposal/consideration process. The table below provides an overview of the current ICTV-approved composition of the order.
Table legend: "*" denotes type species.
Life cycle
The mononegavirus life cycle begins with virion attachment to specific cell-surface receptors, followed by fusion of the virion envelope with cellular membranes and the concomitant release of the virus nucleocapsid into the cytosol. The virus RdRp partially uncoats the nucleocapsid and transcribes the genes into positive-stranded mRNAs, which are then translated into structural and nonstructural proteins. Mononegavirus RdRps bind to a single promoter located at the 3' end of the genome. Transcription either terminates after a gene or continues to the next gene downstream. This means that genes close to the 3' end of the genome are transcribed in the greatest abundance, whereas those toward the 5' end are least likely to be transcribed. The gene order is therefore a simple but effective form of transcriptional regulation. The most abundant protein produced is the nucleoprotein, whose concentration in the cell determines when the RdRp switches from gene transcription to genome replication. Replication results in full-length, positive-stranded antigenomes that are in turn transcribed into negative-stranded virus progeny genome copies. Newly synthesized structural proteins and genomes self-assemble and accumulate near the inside of the cell membrane. Virions bud off from the cell, gaining their envelopes from the cellular membrane they bud from. The mature progeny particles then infect other cells to repeat the cycle.
Paleovirology
Mononegaviruses have a history that dates back several tens of million of years. Mononegavirus "fossils" have been discovered in the form of mononegavirus genes or gene fragments integrated into mammalian genomes. For instance, bornavirus gene "fossils" have been detected in the genomes of bats, fish, hyraxes, marsupials, primates, rodents, ruminants, and elephants. Filovirus gene "fossils" have been detected in the genomes of bats, rodents, shrews, tenrecs, and marsupials. A Midway virus "fossil" was found in the genome of zebrafish. Finally, rhabdovirus "fossils" were found in the genomes of mosquitoes, ticks, and plants.
