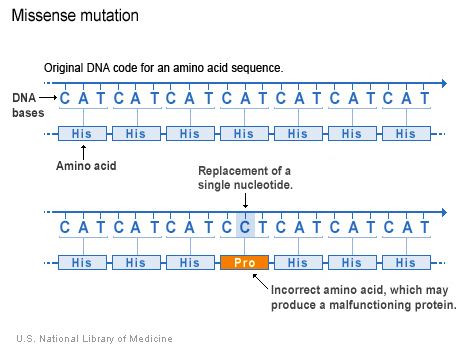
 | ||
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution.
Contents
Substitution of protein from DNA mutations
Missense mutation or substitution refers to a change in one amino acid in a protein, arising from a point mutation in a single nucleotide. Missense mutation is a type of nonsynonymous substitution in a DNA sequence. Another type of nonsynonymous substitution is a nonsense mutation in which a codon is changed to a premature stop codon that results in truncation of the resulting protein. Missense mutations can render the resulting protein nonfunctional, and such mutations are responsible for human diseases such as Epidermolysis bullosa, sickle-cell disease, and SOD1 mediated ALS.
In the most common variant of sickle-cell disease, the 20th nucleotide of the gene for the beta chain of hemoglobin is altered from the codon GAG to GTG. Thus, the 6th amino acid glutamic acid is substituted by valine—notated as an "E6V" mutation—and the protein is sufficiently altered to cause the sickle-cell disease.
Not all missense mutations lead to appreciable protein changes. An amino acid may be replaced by an amino acid of very similar chemical properties, in which case, the protein may still function normally; this is termed a neutral, "quiet", "silent" or conservative mutation. Alternatively, the amino acid substitution could occur in a region of the protein which does not significantly affect the protein secondary structure or function. When an amino acid may be encoded by more than one codon (so-called "degenerate coding") a mutation in a codon may not produce any change in translation; this would be a silent mutation (a type of synonymous substitution, which is not always silent) and not a missense mutation.
Example
DNA: 5' - AAC AGC CTG CGT ACG GCT CTC - 3' 3' - TTG TCG GAC GCA TGC CGA GAG - 5' mRNA: 5' - AAC AGC CUG CGU ACG GCU CUC - 3'Protein: Asn Ser Leu Arg Thr Ala LeuLMNA missense mutation (c.1580G>T) introduced at LMNA gene – position 1580 (nt) in the DNA sequence (CGT) causing the guanine to be replaced with the thymine, yielding CTT in the DNA sequence. This results at the protein level in the replacement of the arginine by the leucine at the position 527. This leads to destruction of salt bridge and structure destabilization. At phenotype level this manifests with overlapping mandibuloacral dysplasia and progeria syndrome.
The resulting transcript and protein product is:
DNA: 5' - AAC AGC CTG CTT ACG GCT CTC - 3' 3' - TTG TCG GAC GAA TGC CGA GAG - 5' mRNA: 5' - AAC AGC CUG CUU ACG GCU CUC - 3'Protein: Asn Ser Leu Leu Thr Ala LeuExperimental analysis
Cancer associated missense mutations can lead to drastic destabilisation of the resulting protein. A novel method to screen for such changes was proposed recently, namely fast parallel proteolysis (FASTpp).
