Possible time of origin 55,000 years ago Ancestor R | Defining mutations 11467, 12308, 12372 | |
 | ||
Descendants U1, U5, U6, U2'3'4'7'8'9 | ||
Haplogroup U is a human mitochondrial DNA haplogroup (mtDNA). Its subclades are widely distributed across Northern Europe, Eastern Europe, Central Asia, South Asia, North Africa, the Horn of Africa, and the Near East.
Contents
Origins
Haplogroup U descends from a woman in the haplogroup R mtDNA branch of the phylogenetic tree, who is estimated to have lived around 55,000 years ago.
Distribution
Haplogroup U is found in 15% of Indian caste and 8% of Indian tribal populations. Haplogroup U is found in approximately 11% of native Europeans and is held as the oldest maternal haplogroup found in that region. In a 2013 study, all but one of the ancient modern human sequences from Europe belonged to maternal haplogroup U, thus confirming previous findings that haplogroup U was the dominant type of Mitochondrial DNA (mtDNA) in Europe before the spread of agriculture into Europe and the presence and the spread of the Indo-Europeans in Western Europe.
Haplogroup U has various subclades numbered U1 to U9. Haplogroup K is a subclade of U8. The old age has led to a wide distribution of the descendant subgroups across Western Eurasia, North Africa, and South Asia. Some subclades of haplogroup U have a more specific geographic range.
Haplogroup U1
Haplogroup U1 is found at very low frequency throughout Europe. It is more often observed in eastern Europe, Anatolia and the Near East. It is also found at low frequencies in India. Haplogroup U1 is a very ancient haplogroup, with an estimated age of about 32,000 years. U1 is found in the Svanetia region at 4.2%. Subclade U1a is found from India to Europe, but is extremely rare among the northern and Atlantic fringes of Europe including the British Isles and Scandinavia. Several examples in Tuscany have been noted. In India, U1a has been found in the Kerala region. U1b has a similar spread but is rarer than U1a. Some examples of U1b have been found among Jewish diaspora. Subclades U1a and U1b appear in equal frequency in eastern Europe.
The U1 subclades are: U1a (with deep-subclades U1a1, U1a1a, U1a1a1, U1a1b) and U1b.
DNA analysis of excavated remains now located at ruins of the Church of St. Augustine in Goa, India revealed the unique mtDNA subclade U1b, which is absent in India, but present in Georgia and surrounding regions. Since the genetic analysis corroborates archaeological and literary evidence, it is believed that the excavated remains belong to Ketevan the Martyr, queen of Georgia.
Haplogroup U1 has been observed among specimens at the mainland cemetery in Kulubnarti, Sudan, which date from the Early Christian period (AD 550-800).
The rare U1 clade is also found among Algerians in Oran (0.83%-1.08%) and the Reguibat tribe of the Sahrawi (0.93%).
Haplogroup U2
Haplogroup U2 is most common in South Asia but is also found in low frequency in Central and West Asia, as well as in Europe as U2e (the European variety of U2 is named U2e). The overall frequency of U2 in South Asia is largely accounted for by the group U2i in India whereas haplogroup U2e, common in Europe, is entirely absent; given that these lineages diverged approximately 50,000-years-ago, these data have been interpreted as indicating very low maternal-line gene-flow between South Asia and Europe throughout this period. Approximately one half of the U mtDNAs in India belong to the Indian-specific branches of haplogroup U2 (U2i: U2a, U2b and U2c). While U2 is typically found in India, it is also present in the Nogais, descendants of various Mongolic and Turkic tribes, who formed the Nogai Horde. Both U2 and U4 are found in the Ket and Nganasan peoples, the indigenous inhabitants of the Yenisei River basin and the Taymyr Peninsula.
The U2 subclades are: U2a, U2b, U2c, U2d, and U2e. With the India-specific subclades U2a, U2b, and U2c collectively referred to as U2i, the Eurasian haplogroup U2d appears to be a sister clade with the Indian haplogroup U2c, while U2e is considered a European-specific subclade but also found in South India.
Haplogroup U2 has been found in the remains of a 30,000-year-old hunter-gatherer from the Kostyonki, Voronezh Oblast in Central-South European Russia., in 4800 to 4000-year-old human remains from a Beaker culture site of the Late Neolithic in Kromsdorf Germany, and in 2,000-year-old human remains from Bøgebjerggård in Southern Denmark. However, haplogroup U2 is rare in present-day Scandinavians. The remains of a 2,000-year-old West Eurasian male of haplogroup U2e1 was found in the Xiongnu Cemetery of Northeast Mongolia.
Haplogroup U3
Haplogroup U3 falls into two subclades:: U3a and U3b.
Coalescence age for U3a is estimated as 18,000 to 26,000-years-ago while the coalescence age for U3b is estimated as 18,000 to 24,000-years-ago. U3a is found in Europe, the Near East, the Caucasus and North Africa. The almost-entirely European distributed subclade, U3a1, dated at 4000 to 7000-years-ago, suggests a relatively recent (late Holocene or later) expansion of these lineages in Europe. There is a minor U3c subclade (derived from U3a), represented by a single Azeri mtDNA from the Caucasus. U3b is widespread across the Middle East and the Caucasus, and it is found especially in Iran, Iraq and Yemen, with a minor European subclade, U3b1b, dated at 2000 to 3000-years-ago. Haplogroup U3 is defined by the HVR1 transition A16343G. It is found at low levels throughout Europe (about 1% of the population), the Near East (about 2.5% of the population), and Central Asia (about 1% of the population). U3 is present in the Svan population from the Svaneti region (about 4.2% of the population) and among Lithuanian Romani, Polish Romani, and Spanish Romani populations (36-56%) consistent with a common migration route from India then out-of-the Balkans for the Lithuanian, Polish, and Spanish Roma.
The U3 clade is also found among Mozabite Berbers (10.59%), as well as Egyptians in the El-Hayez (2.9%) and Gurna oases (2.9%), and Algerians in Oran (1.08%-1.25%). The rare U3a subclade occurs among the Tuareg inhabiting Niger (3.23%).
Haplogroup U3 has been found in some of the 6400-year-old remains (U3a) discovered in the caves at Wadi El‐Makkukh near Jericho associated with the Chalcolithic period. Haplogroup U3 was already present in the West Eurasian gene pool around 6,000-years-ago and probably also its subclade U3a as well.
Haplogroup U4
Haplogroup U4 has its origin in the Upper Paleolithic, dating to approximately 25,000 years ago, and has been implicated in the expansion of modern humans into Europe occurring before the Last Glacial Maximum. U4 is an ancient mitochondrial haplogroup and is relatively rare in modern populations. U4 is found in Europe with highest concentrations in Scandinavia and the Baltic states and is found in the Sami population of the Scandinavian peninsula (although, U5b has a higher representation).
U4 is also associated with the remnants of ancient European hunting-gatherers preserved in the indigenous populations of Siberia. U4 is found in the Nganasan people of the Taymyr Peninsula, in the Mansi (16.3%) an endangered people, and in the Ket people (28.9%) of the Yenisei River.
U4 is also preserved in the Kalash people (current population size 3,700) a unique tribe among the Indo-Aryan peoples of Pakistan where U4 (subclade U4a1) attains its highest frequency of 34%.
The U4 subclades are: U4a, U4b, U4c, and U4d.
Bryan Sykes provided this popular description for haplogroup U4: "The clan of Ulrike (German for Mistress of All) is not among the original 'Seven Daughters of Eve' clans, but with just under 2% of Europeans among its members, it has a claim to being included among the numerically important clans. Ulrike lived about 18,000 years ago in the cold refuges of the Ukraine at the northern limits of human habitation. Though Ulrike's descendants are nowhere common, the clan is found today mainly in the east and north of Europe with particularly high concentrations in Scandinavia and the Baltic states."
Haplogroup U4 is associated with ancient European hunter-gatherers and has been found in 7,200 to 6,000-year-old remains of the Pitted Ware culture in Gotland Sweden and in 4,400 to 3,800-year-old remains from the Damsbo site of the Danish Beaker culture. Remains identified as subclade U4a2 are associated with the Corded Ware culture, which flourished 5200 to 4300 years ago in Eastern and Central Europe and encompassed most of continental northern Europe from the Volga River in the east to the Rhine in the west. Mitochondrial DNA recovered from 3,500 to 3,300-year-old remains at the Bredtoftegård site in Denmark associated with the Nordic Bronze Age include haplogroup U4 with 16179T in its HVR1 indicative of subclade U4c1.
Haplogroup U5
The age of U5 is estimated at 30-50,000 years. Approximately 11% of total Europeans and 10% of European-Americans are in haplogroup U5. In 'The Seven Daughters of Eve', this haplogroup, as well as Haplogroup U in general, is labelled 'Clan Ursula'. Haplogroup U5 is believed to be the oldest single branch of Haplogroup U, hence the sharing of the 'Ursula' designation by both groups.
U5 has been found in human remains dating from the Mesolithic in England, Germany, Lithuania, Poland, Portugal, Russia, Sweden, France and Spain. Neolithic skeletons (~7,000 years old) that were excavated from the Avellaner cave in Catalonia, northeastern Spain included a specimen, which also carried haplogroup U5.
Haplogroup U5 and its subclades U5a and U5b today form the highest population concentrations in the far north, among Sami, Finns, and Estonians. However, it is spread widely at lower levels throughout Europe. This distribution, and the age of the haplogroup, indicate individuals belonging to this clade were part of the initial expansion tracking the retreat of ice sheets from Europe around 10,000 years ago.
Additionally, haplogroup U5 is found in small frequencies and at much lower diversity in the Near East and parts of northern Africa (areas with sizable U6 concentrations), suggesting back-migration of people from Europe toward the south.
Mitochondrial haplogroup U5a has also been associated with HIV infected individuals displaying accelerated progression to AIDS and death.
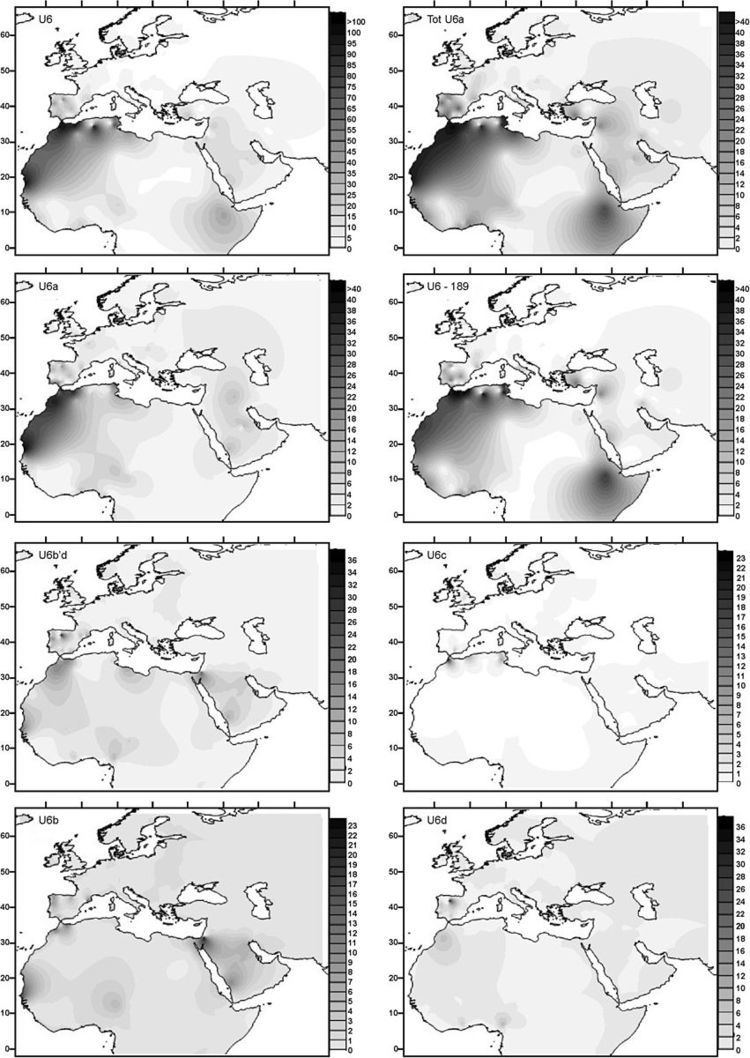
Haplogroup U6
Haplogroup U6 is common (around 10% of the people) in Northwest Africa (with a maximum of 29% in an Algerian Mozabites) and the Canary Islands (18% on average with a peak frequency of 50.1% in La Gomera). It is also found in the Iberian peninsula, where it has the highest diversity (10 out of 19 sublineages are only found in this region and not in Africa), Northeast Africa and occasionally in other locations. U6 is also found at low frequencies in the Chad Basin, including the rare Canarian branch. This suggests that the ancient U6 clade bearers may have inhabited or passed through the Chad Basin on their way westward toward the Canary Islands.
Ancient DNA from a Romanian specimen dated to 35,000 years ago (Peștera Muierilor) has been found to be of the basal U6 haplogroup. The location of this sample suggests a Eurasian origin for the haplogroup, with its dating placing the origin of U6 between 42,000 and 52,000 years ago. U6 is thought to have entered North Africa from the Near East around 30,000 years ago. It has been found among Iberomaurusian specimens dating from the Epipaleolithic at the Taforalt prehistoric site. In spite of the highest diversity of Iberian U6, Maca-Meyer argues for a Near East origin of this clade based on the highest diversity of subclade U6a in that region, where it would have arrived from West Asia, with the Iberian incidence primarily representing migration from the Maghreb and not persistence of a European root population.
According to Hernández et al. 2015 "the estimated entrance of the North African U6 lineages into Iberia at 10 ky correlates well with other L African clades, indicating that U6 and some L lineages moved together from Africa to Iberia in the Early Holocene."
U6 has four main subclades:
Subgroup U6a reflects the first African expansion from the Maghrib returning to the east. Derivative clade U6a1 signals a posterior movement from East Africa back to the Maghrib and the Near East. This migration coincides with the probable Afroasiatic linguistic expansion. U6b and U6c clades, restricted to West Africa, had more localized expansions. U6b probably reached the Iberian Peninsula during the Capsian diffusion in North Africa. Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.
U6a, U6b and U6d share a common basal mutation (16219) that is not present in U6c, whereas U6c has 11 unique mutations. U6b and U6d share a mutation (16311) not shared by U6a, which has three unique mutations.
Haplogroup U6 was named 'Ulla' by Bryan Sykes.
Haplogroup U7
Haplogroup U7 is considered a West Eurasian-specific mtDNA haplogroup, believed to have originated in the Black Sea area approximately 30,000-years-ago. In modern populations, U7 occurs at low frequency in the Caucasus, the western Siberian tribes, West Asia (about 4% in the Near East, while peaking with 10% in Iranians), South Asia (about 12% in Gujarat, the westernmost state of India, while for the whole of India its frequency stays around 2%, and 5% in Pakistan), and the Vedda people of Sri Lanka where it reaches it highest frequency of 13.33% (subclade U7a). One third of the West Eurasian-specific mtDNAs found in India are in haplogroups U7, R2 and W. It is speculated that large-scale immigration carried these mitochondrial haplogroups into India.
The U7 subclades are: U7a (with deep-subclades U7a1, U7a2, U7a2a, U7a2b) and U7b.
Genetic analysis of individuals associated with the Late Hallstatt culture from Baden-Württemberg Germany considered to be examples of Iron Age "princely burials" included haplogroup U7. Haplogroup U7 was reported to have been found in 1200-year-old human remains (dating to around 834), in a woman believed to be from a royal clan who was buried with the Viking Oseberg Ship in Norway. Haplogroup U7 was found in 1000-year-old human remains (dating to around AD 1000-1250) in a Christian cemetery is Kongemarken Denmark. However, U7 is rare among present-day ethnic Scandinavians.
The U7a subclade is especially common among Saudis, constituting around 30% of maternal lineages in the Eastern Province.
Subclade U7a4
The subclade U7a4 point to an origin in the Near East. It peaks among the modern inhabitants of Azerbaijan (26%) and Iranian Azerbaijanis (16-22%), while occurring in the rest of Iran at frequencies from 2-16%, with moderate frequencies of 14-17% in the western parts of Iraq and 1-19% in the Arabian Peninsula. In lower frequencies it is also found in Turkmenistan (2-10%), western parts of Kazakhstan (2-10%), Syria (3-7%), Uzbekistan (2-6%), Central Anatolia (2-4%), South Punjab (1-2%), and Afghanistan (0-2%). U7a4 was also observed in one instance in Tuscany and two other in European Russia. It is presumed that this subclade was brought to Tuscany by the Etruscan civilization. The modern inhabitants of Tuscany are thought to have retained some of this genetic legacy.
Haplogroup U8
Haplogroup K
Haplogroup K makes up a sizeable fraction of European and West Asian mtDNA lineages. It is now known it is actually a subclade of haplogroup U8b'K, and is believed to have first arisen in northeastern Italy. Haplogroup UK shows some evidence of being highly protective against AIDS progression.
Haplogroup U9
Haplogroup U9 is a rare clade in mtDNA phylogeny, characterized only recently in a few populations of Pakistan (Quintana-Murci et al. 2004). Its presence in Ethiopia and Yemen, together with some Indian-specific M lineages in the Yemeni sample, points to gene flow along the coast of the Arabian Sea. Haplogroups U9 and U4 share two common mutations at the root of their phylogeny. It is interesting that, in Pakistan, U9 occurs frequently only among the so-called Makrani population. In this particular population, lineages specific to parts of Eastern Africa occur as frequently as 39%, which suggests that U9 lineages in Pakistan may have an origin from this area (Quintana-Murci et al. 2004). Regardless of which coast of the Arabian Sea may have been the origin of U9, its Ethiopian–southern Arabian–Indus Basin distribution hints that the subclade's diversification from U4 may have occurred in regions far away from the current area of the highest diversity and frequency of haplogroup U4—East Europe and western Siberia.
Tree
This phylogenetic tree of haplogroup U subclades is based on the paper by Mannis van Oven and Manfred Kayser "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation" and subsequent published research.
