In computer science, a suffix tree (also called PAT tree or, in an earlier form, position tree) is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
The construction of such a tree for the string S takes time and space linear in the length of S . Once constructed, several operations can be performed quickly, for instance locating a substring in S , locating a substring if a certain number of mistakes are allowed, locating matches for a regular expression pattern etc. Suffix trees also provide one of the first linear-time solutions for the longest common substring problem. These speedups come at a cost: storing a string's suffix tree typically requires significantly more space than storing the string itself.
The concept was first introduced by Weiner (1973), which Donald Knuth subsequently characterized as "Algorithm of the Year 1973". The construction was greatly simplified by McCreight (1976) , and also by Ukkonen (1995). Ukkonen provided the first online-construction of suffix trees, now known as Ukkonen's algorithm, with running time that matched the then fastest algorithms. These algorithms are all linear-time for a constant-size alphabet, and have worst-case running time of O ( n log n ) in general.
Farach (1997) gave the first suffix tree construction algorithm that is optimal for all alphabets. In particular, this is the first linear-time algorithm for strings drawn from an alphabet of integers in a polynomial range. Farach's algorithm has become the basis for new algorithms for constructing both suffix trees and suffix arrays, for example, in external memory, compressed, succinct, etc.
The suffix tree for the string S of length n is defined as a tree such that:
The tree has exactly n leaves numbered from 1 to n.Except for the root, every internal node has at least two children.Each edge is labeled with a non-empty substring of S.No two edges starting out of a node can have string-labels beginning with the same character.The string obtained by concatenating all the string-labels found on the path from the root to leaf i spells out suffix S[i..n], for i from 1 to n.Since such a tree does not exist for all strings, S is padded with a terminal symbol not seen in the string (usually denoted $). This ensures that no suffix is a prefix of another, and that there will be n leaf nodes, one for each of the n suffixes of S . Since all internal non-root nodes are branching, there can be at most n − 1 such nodes, and n + (n − 1) + 1 = 2n nodes in total (n leaves, n − 1 internal non-root nodes, 1 root).
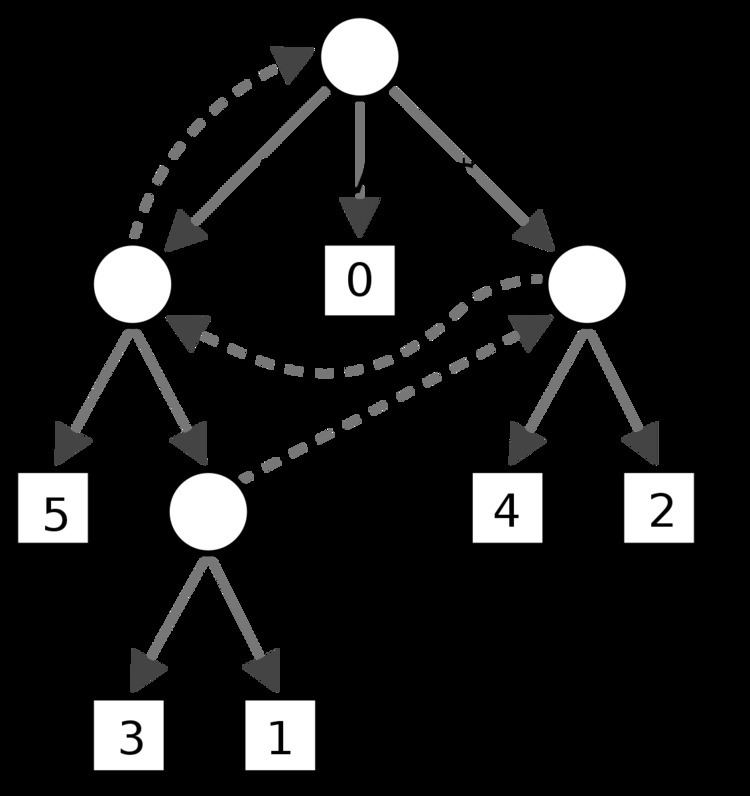
Suffix links are a key feature for older linear-time construction algorithms, although most newer algorithms, which are based on Farach's algorithm, dispense with suffix links. In a complete suffix tree, all internal non-root nodes have a suffix link to another internal node. If the path from the root to a node spells the string χ α , where χ is a single character and α is a string (possibly empty), it has a suffix link to the internal node representing α . See for example the suffix link from the node for ANA to the node for NA in the figure above. Suffix links are also used in some algorithms running on the tree.
A generalized suffix tree is a suffix tree made for a set of words instead of a single word. It represents all suffixes from this set of words. Each word must be terminated by a different termination symbol or word.
A suffix tree for a string S of length n can be built in Θ ( n ) time, if the letters come from an alphabet of integers in a polynomial range (in particular, this is true for constant-sized alphabets). For larger alphabets, the running time is dominated by first sorting the letters to bring them into a range of size O ( n ) ; in general, this takes O ( n log n ) time. The costs below are given under the assumption that the alphabet is constant.
Assume that a suffix tree has been built for the string S of length n , or that a generalised suffix tree has been built for the set of strings D = { S 1 , S 2 , … , S K } of total length n = | n 1 | + | n 2 | + ⋯ + | n K | . You can:
Search for strings:Check if a string P of length m is a substring in O ( m ) time.Find the first occurrence of the patterns P 1 , … , P q of total length m as substrings in O ( m ) time.Find all z occurrences of the patterns P 1 , … , P q of total length m as substrings in O ( m + z ) time.Search for a regular expression P in time expected sublinear in n .Find for each suffix of a pattern P , the length of the longest match between a prefix of P [ i … m ] and a substring in D in Θ ( m ) time. This is termed the matching statistics for P .Find properties of the strings:Find the longest common substrings of the string S i and S j in Θ ( n i + n j ) time.Find all maximal pairs, maximal repeats or supermaximal repeats in Θ ( n + z ) time.Find the Lempel–Ziv decomposition in Θ ( n ) time.Find the longest repeated substrings in Θ ( n ) time.Find the most frequently occurring substrings of a minimum length in Θ ( n ) time.Find the shortest strings from Σ that do not occur in D , in O ( n + z ) time, if there are z such strings.Find the shortest substrings occurring only once in Θ ( n ) time.Find, for each i , the shortest substrings of S i not occurring elsewhere in D in Θ ( n ) time.The suffix tree can be prepared for constant time lowest common ancestor retrieval between nodes in Θ ( n ) time. One can then also:
Find the longest common prefix between the suffixes S i [ p . . n i ] and S j [ q . . n j ] in Θ ( 1 ) .Search for a pattern P of length m with at most k mismatches in O ( k n + z ) time, where z is the number of hits.Find all z maximal palindromes in Θ ( n ) , or Θ ( g n ) time if gaps of length g are allowed, or Θ ( k n ) if k mismatches are allowed.Find all z tandem repeats in O ( n log n + z ) , and k-mismatch tandem repeats in O ( k n log ( n / k ) + z ) .Find the longest common substrings to at least k strings in D for k = 2 , … , K in Θ ( n ) time.Find the longest palindromic substring of a given string (using the generalized suffix tree of the string and its reverse) in linear time.Suffix trees can be used to solve a large number of string problems that occur in text-editing, free-text search, computational biology and other application areas. Primary applications include:
String search, in O(m) complexity, where m is the length of the sub-string (but with initial O(n) time required to build the suffix tree for the string)Finding the longest repeated substringFinding the longest common substringFinding the longest palindrome in a stringSuffix trees are often used in bioinformatics applications, searching for patterns in DNA or protein sequences (which can be viewed as long strings of characters). The ability to search efficiently with mismatches might be considered their greatest strength. Suffix trees are also used in data compression; they can be used to find repeated data, and can be used for the sorting stage of the Burrows–Wheeler transform. Variants of the LZW compression schemes use suffix trees (LZSS). A suffix tree is also used in suffix tree clustering, a data clustering algorithm used in some search engines.
If each node and edge can be represented in Θ ( 1 ) space, the entire tree can be represented in Θ ( n ) space. The total length of all the strings on all of the edges in the tree is O ( n 2 ) , but each edge can be stored as the position and length of a substring of S, giving a total space usage of Θ ( n ) computer words. The worst-case space usage of a suffix tree is seen with a fibonacci word, giving the full 2 n nodes.
An important choice when making a suffix tree implementation is the parent-child relationships between nodes. The most common is using linked lists called sibling lists. Each node has a pointer to its first child, and to the next node in the child list it is a part of. Other implementations with efficient running time properties use hash maps, sorted or unsorted arrays (with array doubling), or balanced search trees. We are interested in:
The cost of finding the child on a given character.The cost of inserting a child.The cost of enlisting all children of a node (divided by the number of children in the table below).Let σ be the size of the alphabet. Then you have the following costs:
Lookup Insertion Traversal Sibling lists / unsorted arrays O ( σ ) Θ ( 1 ) Θ ( 1 ) Bitwise sibling trees O ( log σ ) Θ ( 1 ) Θ ( 1 ) Hash maps Θ ( 1 ) Θ ( 1 ) O ( σ ) Balanced search tree O ( log σ ) O ( log σ ) O ( 1 ) Sorted arrays O ( log σ ) O ( σ ) O ( 1 ) Hash maps + sibling lists O ( 1 ) O ( 1 ) O ( 1 ) The insertion cost is amortised, and that the costs for hashing are given for perfect hashing.
The large amount of information in each edge and node makes the suffix tree very expensive, consuming about 10 to 20 times the memory size of the source text in good implementations. The suffix array reduces this requirement to a factor of 8 (for array including LCP values built within 32-bit address space and 8-bit characters.) This factor depends on the properties and may reach 2 with usage of 4-byte wide characters (needed to contain any symbol in some UNIX-like systems, see wchar_t) on 32-bit systems. Researchers have continued to find smaller indexing structures.
Various parallel algorithms to speed up suffix tree construction have been proposed. Recently, a practical parallel algorithm for suffix tree construction with O ( n ) work (sequential time) and O ( log 2 n ) span has been developed. The algorithm achieves good parallel scalability on shared-memory multicore machines and can index the 3GB human genome in under 3 minutes using a 40-core machine.