Symbol SSB Pfam clan CL0021 PROSITE PDOC00602 | Pfam PF00436 InterPro IPR000424 SCOP 1kaw | |
 | ||
Single-stranded binding proteins (SSBPs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans.
Contents
Function
Single stranded binding proteins prevent premature annealing (binding of complementary DNA sequences), protect the single-stranded DNA from being digested by nucleases, and prevent secondary structure formation, thereby allowing other enzymes to function effectively on the single strand.
Viral SSB
Although the overall picture of human cytomegalovirus (HHV-5) DNA synthesis appears typical of the herpesviruses, some novel features are emerging.
Structure
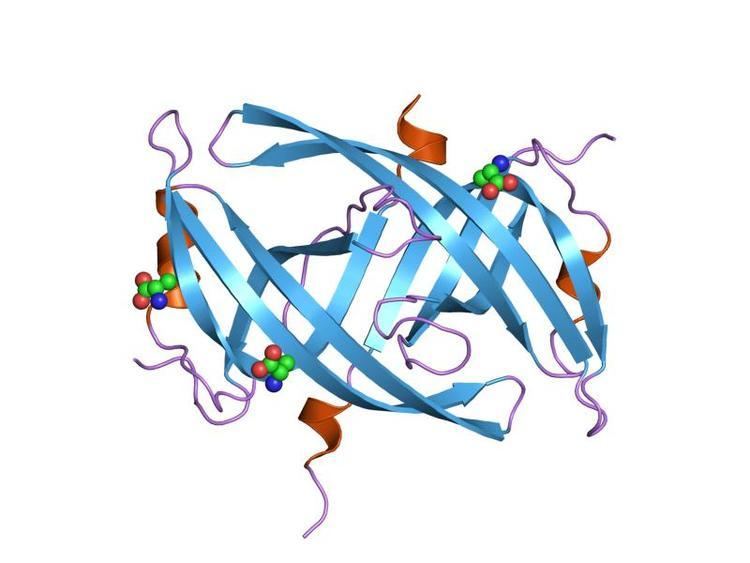
In ICP8, the herpes simplex virus (HSV-1) single-strand DNA-binding protein (ssDNA-binding protein (SSB)), The head consists of the eight alpha helices. The front side of the neck region consists of a five-stranded beta-sheet and two alpha helices, whereas the back side is a three-stranded beta-sheet The shoulder part of the N-terminal domain contains an alpha-helical and beta-sheet region. The herpes simplex virus (HSV-1) SSB, ICP8, is a nuclear protein that, along other replication proteins is required for viral DNA replication during lytic infection.
Mechanism
Six herpes virus-group-common genes encode proteins that likely constitute the replication fork machinery, including a two-subunit DNA polymerase, a Helicase-primase complex and a single-stranded DNA-binding protein. The human herpesvirus 1 (HHV-1) single-strand DNA-binding protein ICP8 is a 128kDa zinc metalloprotein. Photoaffinity labeling has shown that the region encompassing amino acid residues 368-902 contains the single-strand DNA-binding site of ICP8. The HHHV-1 UL5, UL8, and UL52 genes encode an essential heterotrimeric DNA helicase-primase that is responsible for concomitant DNA unwinding and primer synthesis at the viral DNA replication fork. ICP8 may stimulate DNA unwinding and enable bypass of cisplatin damaged DNA by recruiting the helicase-primase to the DNA.
Bacterial SSB
In molecular biology, SSB protein domains in bacteria are important maintaining DNA metabolism, more specifically DNA replication, repair and recombination. It has a structure of three beta-strands to a single six-stranded beta-sheet to form a dimer.
Eukaryotic replication protein A
Replication protein A is the functional equivalent of SSB in the nucleus of eukaryotic cells, though there is no sequence homology.
Eukaryotic mitochondrial SSB
The mitochondria of eukaryotic cells contain their own single stranded DNA binding protein. Human mitochondrial SSB (mtSSB) binds to single-stranded mitochondrial DNA as a tetramer and has sequence similarity to bacterial SSB. Human mtSSB is encoded by the SSBP1 gene. In yeast, it is encoded by the RIM1 gene.
