Symbol SAG InterPro IPR007226 SUPERFAMILY 1kzq | Pfam PF04092 SCOP 1kzq Pfam structures | |
 | ||
In molecular biology, the SAG1 protein domain is an example of a group of glycosylphosphatidylinositol (GPI)-linked proteins named SRSs (SAG1 related sequence). SAG1 is found on the surface of a protozoan parasite Toxoplasma gondii. This parasite infects almost any warm-blooded vertebrate. The surface of T. gondii is coated with a family of developmentally regulated glycosylphosphatidylinositol (GPI)-linked proteins (SRSs), of which SAG1 is the prototypic member.
Contents
Function
SAG1 and the rest of the SRS protein family mediate cell adhesion to the host cell. They also recognise herapin sulphate proteoglycans, which are part of the Extracellular matrix.
Structure
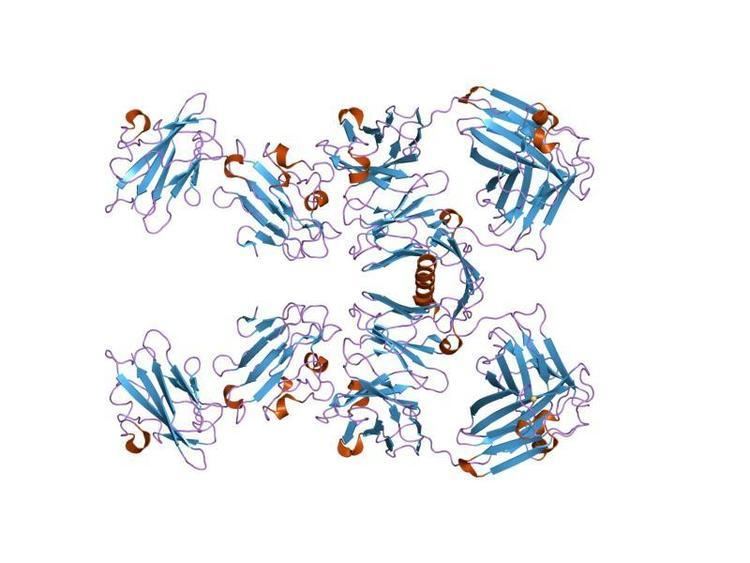
The structure of the immunodominant SAG1 antigen reveals a homodimeric configuration. This family of surface antigens is found in other apicomplexans. This particular antigen contains many cysteine residues which lead to disulphide bridge formation.
N-terminal domain
This domain has a sequence length of at least 130 amino acids which fold to give a beta sheet sandwich.
C terminal domain
This protein domain contains 120 amino acids.
SRS fold
There is a fold within this structure which is conserved amongst SRS proteins, and therefore named the SRS fold. The role of this fold is to aid the binding to cells, and promote infection within the host organism. The structure of this fold is analogous to the topology of the cupredoxin, azurin, a form of copper binding protein.
