Family Acholeplasmataceae Rank Genus | ||
 | ||
Similar Mollicutes, Candidatus Phytoplasma solani, Leafhopper, Scaphoideus titanus, Spiroplasma | ||
Phytoplasmas in plants foodskey
Phytoplasmas are obligate bacterial parasites of plant phloem tissue and of the insect vectors that are involved in their plant-to-plant transmission. Phytoplasmas were discovered in 1967 by Japanese scientists who termed them mycoplasma-like organisms or MLOs. Since their discovery, phytoplasmas have resisted all attempts at in vitro culture in any cell-free medium; routine cultivation in an artificial medium thus remains a major challenge. Although phytoplasmas have recently been reported to be grown in a specific artificial medium, no repetition have been reported. Phytoplasmas are characterized by the lack of a cell wall, a pleiomorphic or filamentous shape, a diameter normally less than 1 μm, and a very small genome.
Contents
- Phytoplasmas in plants foodskey
- nippon gene lamp dried premix phytoplasma universal detection kit
- History
- Morphology
- Symptoms
- Effector virulence proteins
- Movement between plants
- Movement within plants
- Detection and diagnosis
- Control
- Genetics
- Taxonomy
- References
Phytoplasmas are pathogens of agriculturally important plants, including coconut, sugarcane, and sandalwood, in which they cause a wide variety of symptoms ranging from mild yellowing to death. Phytoplasmas are most prevalent in tropical and subtropical regions. They are transmitted from plant to plant by vectors (normally sap-sucking insects such as leafhoppers) in which they both survive and replicate.
nippon gene lamp dried premix phytoplasma universal detection kit
History
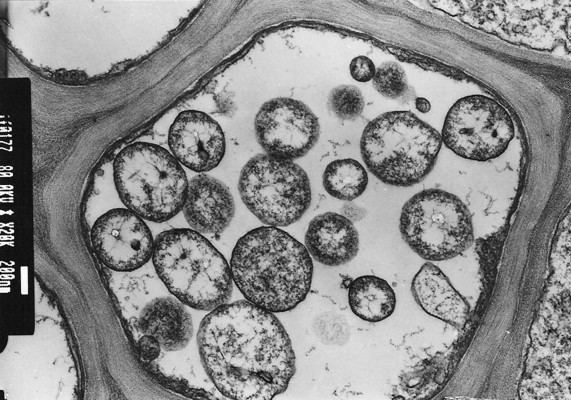
References to diseases now known to be caused by phytoplasmas can be found as far back as 1603 (mulberry dwarf disease in Japan.) Such diseases were originally thought to be caused by viruses, which, like phytoplasmas, require insect vectors, and cannot be cultured. Viral and phytoplasmic infections share some symptoms. In 1967, phytoplasmas were discovered in ultrathin sections of plant phloem tissue and were termed Mycoplasma-like organisms (MLOs) due to their physiological resemblance The organisms were renamed phytoplasmas in 1994, at the 10th Congress of the International Organization for Mycoplasmology.
Morphology

Phytoplasmas are Mollicutes, which are bound by a triple-layered membrane, rather than a cell wall. The phytoplasma cell membranes studied to date usually contain a single immunodominant protein of unknown function that constitutes most of the protein in the membrane. A typical phytoplasma is pleiomorphic or filamentous in shape and is less than 1 μm in diameter. Like other prokaryotes, phytoplasmic DNA is distributed throughout the cytoplasm, instead of being concentrated in a nucleus.
Symptoms
Phytoplasmas can infect and cause various symptoms in more than 700 plant species. One characteristic symptom is abnormal floral organ development including phyllody, (i.e., the production of leaf-like structures in place of flowers) and virescence (i.e., the development of green flowers attributable to a loss of pigment by petal cells). Phytoplasma-harboring flowering plants may nevertheless be sterile. The expression of genes involved in maintaining the apical meristem or in the development of floral organs is altered in the morphologically affected floral organs of phytoplasma-infected plants.
A phytoplasma infection often triggers leaf yellowing, probably due to the presence of phytoplasma cells in phloem, which can affect phloem function and carbohydrate transport, inhibit chlorophyll biosynthesis, and trigger chlorophyll breakdown. These symptoms may be attributable to stress caused by the infection rather than a specific pathogenetic process.
Many phytoplasma-infected plants develop a bushy or "witches' broom" appearance due to changes in their normal growth patterns. Most plants exhibit apical dominance but infection can trigger the proliferation of auxiliary (side) shoots and a reduction in internode size. Such symptoms are actually useful in the commercial production of poinsettias. Infection triggers more axillary shoot production; the poinsettia plants thus produce more than a single flower.
Effector (virulence) proteins
Many plant pathogens produce virulence factors (i.e., effectors) that modulate or interfere with normal host processes to the benefit of the pathogens. In 2009, a secreted protein, termed “tengu-su inducer” (TENGU), was identified from a phytoplasma causing yellowing of onions; this was the first phytoplasmal virulence factor to be described. TENGU induces characteristic symptoms (termed “tengu-su”), including witches’ broom and dwarfism. Transgenic expression of TENGU in Arabidopsis plants induced sterility in male and female flowers. TENGU contains a signal peptide at its N-terminus; after cleavage, the mature protein is only 38 amino acids in length. Although phytoplasmas are restricted to phloem, TENGU is transported from phloem to other cells, including those of the apical and axillary meristems. TENGU was suggested to inhibit both auxin- and jasmonic acid-related pathways, thereby affecting plant development. Surprisingly, the N-terminal 11 amino acid region of the mature protein triggers symptom development in Nicotiana benthamiana plants. TENGU undergoes proteolytic processing by a plant serine protease in vivo, suggesting that the N-terminal peptide (i.e., the 11 amino acid fragment) alone induces the observed symptoms. TENGU homologs have been identified in AY-group phytoplasmas. All such homologs undergo processing and can induce symptoms, suggesting that the symptom-inducing mechanism is conserved among TENGU homologs.
TCP transcription factors normally regulate plant development, controlling the expression of lipoxygenase genes required for jasmonate biosynthesis. Jasmonate levels are decreased in phytoplasma-infected Arabidopsis plants and plants that transgenically express SAP11, an effector of AY-WB phytoplasmas. The downregulation of jasmonate production is beneficial to phytoplasmas because jasmonate is involved in plant defenses against herbivorous insects such as leafhoppers. Leafhoppers lay increased numbers of eggs on AY-WB-infected plants, at least in part because of SAP11 production. For example, the leafhopper Macrosteles quadrilineatus laid 30% more eggs on plants that expressing SAP11 transgenically than control plants, and 60% more eggs on plants infected with AY-WB. Phytoplasmas cannot survive in the external environment and are dependent upon insects such as leafhoppers for transmission to new (healthy) plants. Thus, by compromising jasmonate production, SAP11 'encourages' leafhoppers to lay more eggs on phytoplasma-infected plants, thereby ensuring that newly hatched leafhopper nymphs feed upon infected plants to become phytoplasma vectors.
A phytoplasma effector protein, SAP54, has been shown to induce virescence and Phyllody when expressed in plants. SAP54 homologs have been identified in various phytoplasma species. Two SAP54 homologs, PHYL1 of the onion yellows phytoplasma and PHYL1PnWB of the peanut witches’ broom phytoplasma, induce phyllody-like floral abnormalities. These results suggest that PHYL1, SAP54, and their homologs form a phyllody-inducing gene family, the members of which are termed phyllogens. MADS-box transcription factors (MTFs) of the ABCE model play critical roles in floral organ development in Arabidopsis. Phyllogens interact directly with class A and class E MTFs, inducing protein degradation in a ubiquitin/proteasome-dependent manner. The accumulation of mRNAs encoding class B MTFs, the transcription of which is positively regulated by class A and class E MTFs, is drastically decreased in Arabidopsis constitutively expressing PHYL1. Phyllogens induce abnormal floral organ development by inhibiting the functions of these MTFs.
Movement between plants
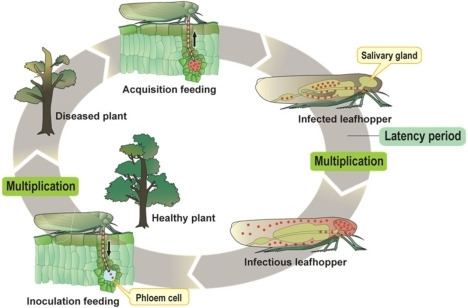
Phytoplasmas are spread principally by insects of the families Cicadellidae (leafhoppers), Fulgoridae (planthoppers), and Psyllidae (jumping plant lice) , which feed on the phloem of infected plants, ingesting phytoplasmas and transmitting them to the next plant on which they feed. Thus, the host range of phytoplasmas is strongly dependent upon that of the insect vector. Phytoplasmas contain a major antigenic protein constituting most of the cell surface protein. This protein associates with insect microfilament complexes and is believed to control insect-phytoplasma interactions. Phytoplasmas can overwinter in insect vectors or perennial plants. Phytoplasmas can have varying effects on their insect hosts; examples of both reduced and increased fitness have been noted.
Phytoplasmas enter the insect body through the stylet, pass through the intestine, and then move to the hemolymph and colonize the salivary glands: the entire process that can take up to 3 weeks. Once established in an insect host, phytoplasmas are found in most major organs. The time between ingestion by the insect and attainment of an infectious titer in the salivary glands is termed the latency period.
Phytoplasmas can also be spread via dodders (Cuscuta) or by vegetative propagation such as the grafting of infected plant tissue onto a healthy plant.
Movement within plants
Phytoplasmas move within phloem from a source to a sink, and can pass through sieve tubes. However, as phytoplasmas spread more slowly than solutes, and for other reasons, passive translocation within plants is thought to be unimportant
Detection and diagnosis
Before the molecular era, the diagnosis of phytoplasma-caused diseases was difficult because the organisms could not be cultured. Thus, classical diagnostic techniques, including symptom observation were used. Ultrathin sections of phloem tissue from platns with suspected phytoplasma-infections were also studied. The empirical use of antibiotics such as tetracycline was additionally employed.
Molecular diagnostic techniques for phytoplasma detection began to emerge in the 1980s and included enzyme-linked immunosorbent assay (ELISA)-based methods. In the early 1990s, polymerase chain reaction (PCR)-based techniques were developed: these are far more sensitive than ELISAs, and restriction fragment length polymorphism (RFLP) analysis allowed the accurate identification of various phytoplasma strains and species.
More recent techniques allow infection levels to be assessed. Both quantitative PCR and bioimaging can effectively quantify phytoplasma titers within plant. In addition, loop-mediated isothermal amplification (a sensitive, simple, and rapid diagnostic method) is now available as a commercial kit allowing all known phytoplasma species to be detected in about 1 h, including the DNA extraction step.
Control
Phytoplasmas are normally controlled by the breeding and planting of disease-resistant crop varieties (perhaps the most economically viable option) and by the control of insect vectors.
Tissue culture can be used to produce healthy clones of phytoplasma-infected plants. Cryotherapy (i.e., the freezing of plant samples in liquid nitrogen) prior to tissue culture increases the probability of producing healthy plants in this manner.
Plantibodies targeting phytoplasmas have also been developed.
Tetracyclines are bacteriostatic to phytoplasmas. However, disease symptoms reappear in the absence of continuous antibiotic application. Thus, tetracycline is not a viable agricultural control agent, but it is used to protect ornamental coconut trees.
Genetics
The genomes of four phytoplasmas have been sequenced: "onion yellows", "aster yellows witches' broom" (Candidatus [Ca] Phytoplasma asteris), Ca. Phytoplasma australiense, and Ca. Phytoplasma Mali. Phytoplasmas have very small genomes, with extremely small amount of G and C nucleotides (sometimes as little as 23%, which is thought to be the lower threshold for a viable genome). In fact, the Bermuda grass white-leaf phytoplasma has a genome size of only 530 kb, one of the smallest known genomes of all living organisms. The larger phytoplasma genomes are around 1350 kb in size. The small genome size of phytoplasma is attributable to reductive evolution from Bacillus/Clostridium ancestors. Phytoplasmas have lost ≥75% of their original genes, and can thus no longer survive outside of insects or plant phloem. Some phytoplasmas contain extrachromosomal DNA such as plasmids.
Despite their small genomes, many predicted phytoplasma genes are present in multiple copies. Phytoplasmas lack many genes encoding standard metabolic functions and have no functioning homologous recombination pathway, but they do have a sec transport pathway. Many phytoplasmas contain two rRNA operons. Unlike other Mollicutes, the triplet code of UGA is used as a stop codon in phytoplasmas.
Phytoplasma genomes contain large numbers of transposons and insertion sequences and also contain a unique family of repetitive extragenic palindromes termed PhREPS for which no role is known. However, it is theorized that the stem-loop structures in PhREPS play a role in transcription termination or genome stability.
Taxonomy
Phytoplasmas belong to the monophyletic order Acholeplasmatales. In 1992, the Subcommittee on the taxonomy of Mollicutes proposed the use of "Phytoplasma" rather than "MLO" "for reference to the phytopathogenic mollicutes". In 2004, the generic name phytoplasma was adopted and is currently of Ca. status (used for Bacteria that cannot be cultured). Phytoplasma taxonomy is complicated because the organisms cannot be cultured; methods normally used to classify prokaryotes are thus not available. Phytoplasma taxonomic groups are based on differences in fragment sizes produced by restriction digests of 16S rRNA gene sequences (RFLPs) or by comparisons of DNA sequences from 16s/23s spacer regions. The actual number of taxonomic groups remains unclear; recent work on computer-simulated restriction digests of the 16Sr gene suggested up to 28 groups, whereas others have proposed fewer groups, but more subgroups. Each group includes at least one Ca. Phytoplasma species, characterized by distinctive biological, phytopathological, and genetic properties.
