Ancestor CF | ||
 | ||
Possible time of origin 57,500–62,500;(Raghavan 2014);45,000–55,700 BP (Karafet 2008);43,000–56,800 BP (Hammer & Zegura 2002). Defining mutations M89/PF2746, L132.1, M213/P137/Page38, M235/Page80, P14, P133, P134, P135, P136, P138, P139, P140, P141,P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187, P316 | ||
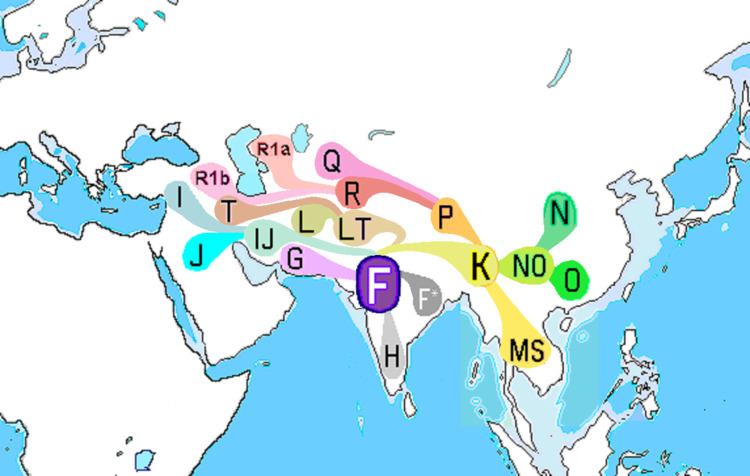
Haplogroup F, also known as F-M89 and previously as Haplogroup FT is a very common Y-chromosome haplogroup. The clade and its subclades constitute over 90% of paternal lineages outside of Africa. It is primarily found throughout South Asia, Southeast Asia and parts of East Asia.
Contents
- Origins
- Distribution
- FxGHIJK
- F M89
- F1 P91
- F2 M427
- F3 M481
- Haplogroup GHIJK
- Phylogenetics
- Phylogenetic trees
- The Genomic Research Center draft tree
- YCCISOGG tree
- Phylogenetic history
- References
The vast majority of individual males with F-M89 fall into its direct descendant Haplogroup GHIJK (F1329/M3658/PF2622/YSC0001299). in addition to GHIJK, haplogroup F has three other immediate descendant subclades: F1 (P91/P104), F2 (M427/M428), and F3 (M481). These three, with F* (M89*), constitute the paragroup F(xGHIJK).
Haplogroup GHIJK branches subsequently into two direct descendants: G (M201/PF2957) and HIJK (F929/M578/PF3494/S6397). HIJK in turn splits into H (L901/M2939) and IJK (F-L15). The descendants of Haplogroup IJK include haplogroups I, J, K, and, ultimately, several major haplogroups descended from Haplogroup K, namely: haplogroups M, N, O, P, Q, R, S, L, and T.
Origins
This megahaplogroup contains mainly lineages that are not typically found in sub-Saharan Africa, suggesting that its immediate ancestor haplogroup CF may have been carried out of Africa very early in the modern human diaspora, suggesting that F-M89 appeared 38,700-55,700 years ago, probably in South Asia.
The descendants of Basal F-M89* are found mostly in populations of South Asia, Southeast Asia and parts of East Asia.
Other sources suggest that this ancient haplogroup may have first appeared in North Africa, the Levant, or the Arabian Peninsula, as much as 50,000 years ago (50,300±6500). It is sometimes believed to represent a "second-wave" of expansion out of Africa. However, the location of this lineage's first expansion and rise to prevalence appears to have been in South Asia or somewhere close to it within the extended Middle East. Most of F's descendant haplogroups appear to radiate outward from South Asia and/or the Middle East.
Some lineages derived from Haplogroup F-M89 appear to have migrated into Africa from Southwest Asia, during prehistory. Y-chromosome haplogroups associated with this hypothetical "Back to Africa" migration include J, R1b, and T.
Distribution
The vast majority of living individuals carrying F-M89 belong to subclades of GHIJK. By comparison, cases of F xG,H,I,J,K – that is, either basal F* (M89) or the primary subclades F1 (P91; P104), F2 (M427; M428) and F3 (M481) – are relatively rare worldwide.
F xG,H,I,J,K
A lack of precise, high resolution testing in the past makes it difficult to discuss F*, F1, F2* and F3* separately. ISOGG states that F xG,H,I,J,K has not been well studied, occurs "infrequently" in modern populations and peaks in South Asia, especially Sri Lanka. It also appears to have long been present in South East Asia. However, the possibility of misidentification is considered to be relatively high and some cases may in fact belong to misidentified subclades of Haplogroup GHIJK. This was, for instance, the case with the subclade Haplogroup H2, (H-M96), which was originally named "F3", i.e – a name that has since been reassigned to F-M481.
F* (F xF1,F2,F3) has been reported among 10% of males in Sri Lanka, 5.2% of males across India (including up to 10% of males in South India), 5% in Pakistan, as well as lower levels among the Tamang people (Nepal), and in Iran. Men originating in Indonesia have also been reported to carry F xG,H,I,J,K – especially F* – at relatively significant levels. It has been reported at rates of 4-5% in Sulawesi and Lembata. One study, which did not comprehensively screen for other subclades of F-M89 (including some subclades of GHIJK), found that Indonesian men with the SNP P14/PF2704 (which is equivalent to M89), comprise 1.8% of men in West Timor, 1.5% of Flores 5.4% of Lembata 2.3% of Sulawesi and 0.2% in Sumatra. F1 (P91), F2 (M427) and F3 (M481; previously F5) are all highly rare and virtually exclusive to regions/ethnic minorities in Sri Lanka, India, Nepal, South China, Thailand, Burma, and Vietnam.
In Central Asia, examples of F xG,H,I,J,K have been reported, in individuals from Turkmenistan and Uzbekistan.
There is also evidence of significant westward Paleolithic back-migration of F xG,H,I,J,K from South Asia, to Iran, Arabia and Northeast Africa, as well as South-East Europe. An individual known to scholars as "Oase 1", who lived circa 37,800 years BP in Romania falls into F xG,H,I,J,K.
Neolithic migration into Europe from Southwest Asia, by first wave of farmers in Europe has been put forward as the source of F(xGHIJK) and G2a found in European Neolithic remains, dating from circa 4000 BCE. These remains, according to Herrerra et al. (2012) showed a "greater genetic similarity" to "individuals from the modern Near East" than to modern Europeans. F xG,H,I,J,K has also apparently been found in Bronze Age remains from Europe:
Some cases reported amongst modern populations of Europeans, Native Americans and Pacific Islanders may be due to migration and admixture of F xG,H,I,J,K as a result of contact with South and South East Asia during the early modern era (16th–19th Century). Such examples include:
F* (M89*)
Basal F-M89* has been reported among 5.2% of males in India. A regional breakdown was provided by Chiaroni et al (2009): 10% in South India; 8% in Central India; about 1.0% in North India and Western India, as well as 5% in Pakistan; 10% in Sri Lanka; >4% among the Tamang people of Nepal; 2% in Borneo and Java; 4-5% in Sulawesi and Lembata.
In Iran, 2.3% of Bandari males from Hormozgan Province have been found to carry basal F-M89.
Haplogroup F-M89 has also been observed in Northeast Africa among two Christian period individuals, who were excavated on the Nile's Fourth Cataract and on Meroe Island.
F1 (P91)
This subclade is defined by the SNP P91. It is most common in Sri Lanka.
F2 (M427)
F2 Y-chromosomes have been reported among minorities from the borderlands of South China (Yunnan and Guizhou), Thailand, Burma, and Vietnam, namely the Yi and Kucong or Lahu Shi ("Yellow Lahu"), a subgroup of the Lahu.
F3 (M481)
The newly defined and rare subclade F3 (M481; previously F5) has been found in India and Nepal, among the Tharu people and in Andhra Pradesh. F-M481 should not be confused with Haplogroup H2 (L279, L281, L284, L285, L286, M282, P96), which was previously misclassified under F-M89, as "F3".
Haplogroup GHIJK
Basal GHIJK has never been found, either in living males or ancient remains.
Subclades – including some major haplogroups – are widespread in modern populations of the Caucasus, Middle East, South Asia, Europe South East Asia, Pacific Islands and Native Americans.
Phylogenetics
In Y-chromosome phylogenetics, subclades are the branches of haplogroups. These subclades are also defined by single nucleotide polymorphisms (SNPs) or unique event polymorphisms (UEPs).
Phylogenetic trees
There are several confirmed and proposed phylogenetic trees available for haplogroup F-M89. The scientifically accepted one is the Y-Chromosome Consortium (YCC) one published in Karafet 2008 and subsequently updated. A draft tree that shows emerging science is provided by Thomas Krahn at the Genomic Research Center in Houston, Texas. The International Society of Genetic Genealogy (ISOGG) also provides an amateur tree.
The Genomic Research Center draft tree
he Genomic Research Center's draft tree for haplogroup F-M89 is as follows. (Only the first three levels of subclades are shown.)
YCC/ISOGG tree
This is the official scientific tree produced by the Y-Chromosome Consortium (YCC). The last major update was in 2008. Subsequent updates have been quarterly and biannual. The current version is a revision of the 2010 update.
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
