Genus Francisella Rank Species Higher classification Francisella | Scientific name Francisella tularensis Phylum Proteobacteria | |
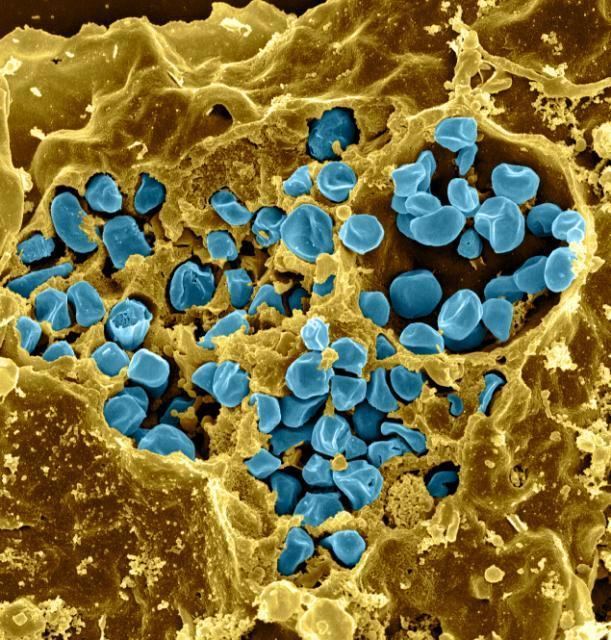 | ||
Similar Francisella, Bacteria, Coxiella, Brucella, Rickettsia | ||
Tularemia and francisella tularensis
Francisella tularensis is a pathogenic species of Gram-negative, rod-shaped coccobacillus, an aerobe bacterium. It is non-spore forming, non-motile and the causative agent of tularemia, the pneumonic form of which is often lethal without treatment. It is a fastidious, facultative intracellular bacterium which requires cysteine for growth. Due to its low infectious dose, ease of spread by aerosol, and high virulence, F. tularensis is classified as a Tier 1 Select Agent by the U.S. government, along with other potential agents of bioterrorism such as Yersinia pestis, Bacillus anthracis and Ebola virus.
Contents
- Tularemia and francisella tularensis
- Subspecies
- Pathogenesis
- Lifecycle
- Virulence factors
- Genetics
- Phylogenetics
- Use as a biological weapon
- Diagnosis treatment and prevention
- Genomics
- References
Subspecies
This species was discovered in ground squirrels in Tulare County, California, in 1911; Bacterium tularense was soon isolated by George Walter McCoy of the US Plague Lab in San Francisco and reported in 1912. In 1922, Dr. Edward Francis (1872–1957), a Physician and Medical Researcher from Ohio, discovered that the "Bacterium Tularense" was the causative agent for tularemia, after studying several cases of his patients having symptoms of the said disease. Later, Bacterium Tularense would become known as "Francisella Tularensis", in honor of the discovery of Dr. Francis. Four subspecies (biovars) of F. tularensis have been classified.
- The subspecies F. t. tularensis (or type A) is found predominantly in North America, is the most virulent of the four known subspecies, and is associated with lethal pulmonary infections. This includes the primary type A laboratory strain, SCHUS4.
- Subspecies F. t. holarctica (also known as biovar F. t. palearctica or type B) is found predominantly in Europe and Asia, but rarely leads to fatal disease. An attenuated live vaccine strain of subspecies F. t. holarctica has been described, though it is not yet fully licensed by the FDA as a vaccine. This subspecies lacks the citrulline ureidase activity and ability to produce acid from glucose of biovar F. t. palearctica.
- Subspecies F. t. novicida (previously classified as F. novicida) was characterized as a relatively nonvirulent strain; only two tularemia cases in North America have been attributed to F. t. novicida and these were only in severely immunocompromised individuals.
- Subspecies F. t. mediasiatica, is found primarily in central Asia; little is currently known about this subspecies or its ability to infect humans.
Pathogenesis
F. tularensis has been reported in birds, reptiles, fish, invertebrates, and mammals including humans. Despite this, no case of tularemia has been shown to be initiated by human-to-human transmission. Rather, tularemia is caused by contact with infected animals or vectors such as ticks, mosquitos, and deer flies. Reservoir hosts of importance can include lagomorphs, rodents, galliform birds, and deer.
Infection with F. tularensis can occur by several routes. The most common occurs via skin contact, yielding an ulceroglandular form of the disease. Inhalation of bacteria - particularly biovar F. t. tularensis, leads to the potentially lethal pneumonic tularemia. While the pulmonary and ulceroglandular forms of tularemia are more common, other routes of inoculation have been described and include oropharyngeal infection due to consumption of contaminated food and conjunctival infection due to inoculation at the eye.
F. tularensis is capable of surviving outside of a mammalian host for weeks at a time and has been found in water, grassland, and haystacks. Aerosols containing the bacteria may be generated by disturbing carcasses due to brush cutting or lawn mowing; as a result, tularemia has been referred to as "lawnmower disease". Recent epidemiological studies have shown a positive correlation between occupations involving the above activities and infection with F. tularensis.
Lifecycle
F. tularensis is a facultative intracellular bacterium that is capable of infecting most cell types, but primarily infects macrophages in the host organism. Entry into the macrophage occurs by phagocytosis and the bacterium is sequestered from the interior of the infected cell by a phagosome. F. tularensis then breaks out of this phagosome into the cytosol and rapidly proliferates. Eventually, the infected cell undergoes apoptosis, and the progeny bacteria are released to initiate new rounds of infection.
Virulence factors
The virulence mechanisms for F. tularensis have not been well characterized. Like other intracellular bacteria that break out of phagosomal compartments to replicate in the cytosol, F. tularensis strains produce different hemolytic agents, which may facilitate degradation of the phagosome. A hemolysin activity, named NlyA, with immunological reactivity to Escherichia coli anti-HlyA antibody, was identified in biovar F. t. novicida. Acid phosphatase AcpA has been found in other bacteria to act as a hemolysin, whereas in Francisella, its role as a virulence factor is under vigorous debate.
While F. tularensis does not contain virulence secretion systems typical of some better-characterized pathogenic bacteria, it does contain a number of ATP-binding cassette (ABC) proteins that may be linked to the secretion of virulence factors. F. tularensis uses type IV pili to bind to the exterior of a host cell and thus become phagocytosed. Mutant strains lacking pili show severely attenuated pathogenicity.
The expression of a 23-kD protein known as IglC is required for F. tularensis phagosomal breakout and intracellular replication; in its absence, mutant F. tularensis cells die and are degraded by the macrophage. This protein is located in a putative pathogenicity island regulated by the transcription factor MglA.
F. tularensis, in vitro, downregulates the immune response of infected cells, a tactic used by a significant number of pathogenic organisms to ensure their replication is (albeit briefly) unhindered by the host immune system by blocking the warning signals from the infected cells. This downmodulation of the immune response requires the IglC protein, though again the contributions of IglC and other genes are unclear. Several other putative virulence genes exist, but have yet to be characterized for function in F. tularensis pathogenicity.
Genetics
Like many other bacteria, F. tularensis undergoes asexual replication. Bacteria divide into two daughter cells, each of which contains identical genetic information. Genetic variation may be introduced by mutation or horizontal gene transfer.
The genome of F. t. tularensis strain SCHU4 has been sequenced. The studies resulting from the sequencing suggest a number of gene-coding regions in the F. tularensis genome are disrupted by mutations, thus create blocks in a number of metabolic and synthetic pathways required for survival. This indicates F. tularensis has evolved to depend on the host organism for certain nutrients and other processes ordinarily taken care of by these disrupted genes.
The F. tularensis genome contains unusual transposon-like elements resembling counterparts that normally are found in eukaryotic organisms.
Phylogenetics
Much of the known global genetic diversity of F. t. holarctica is present in Sweden. This suggests this subspecies originated in Scandinavia and spread from there to the rest of Eurosiberia.
Use as a biological weapon
When the U.S. biological warfare program ended in 1969, F. tularensis was one of seven standardized biological weapons it had developed.
Diagnosis, treatment, and prevention
Infection by F. tularensis is diagnosed by clinician based on symptoms and patient history, imaging, and laboratory studies.
Tularemia is treated with antibiotics, such as aminoglycosides, tetracyclines, or fluoroquinolones. 15 proteins were suggested which could facillate drug and vaccine design pipline.
