Genus Rhodospirillum Rank Species | Scientific name Rhodospirillum rubrum Higher classification Rhodospirillum | |
 | ||
Similar Rhodospirillum, Bacteria, Rhodopseudomonas, Rhodobacter sphaeroides, Rhodobacter | ||
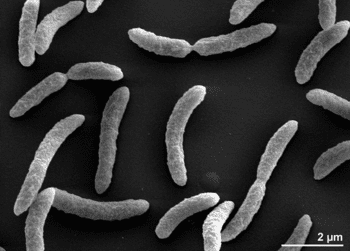
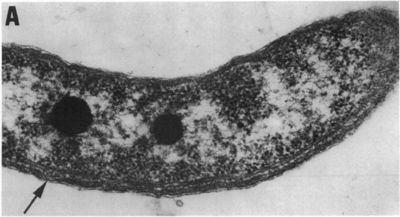
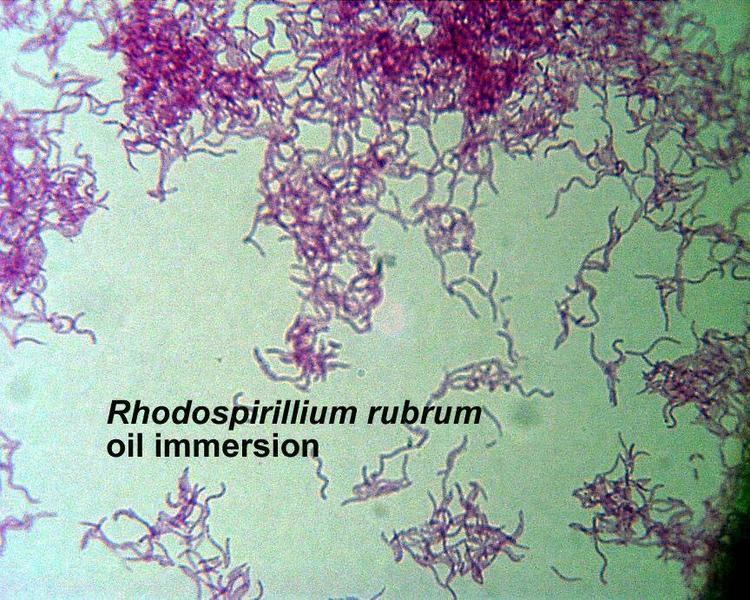
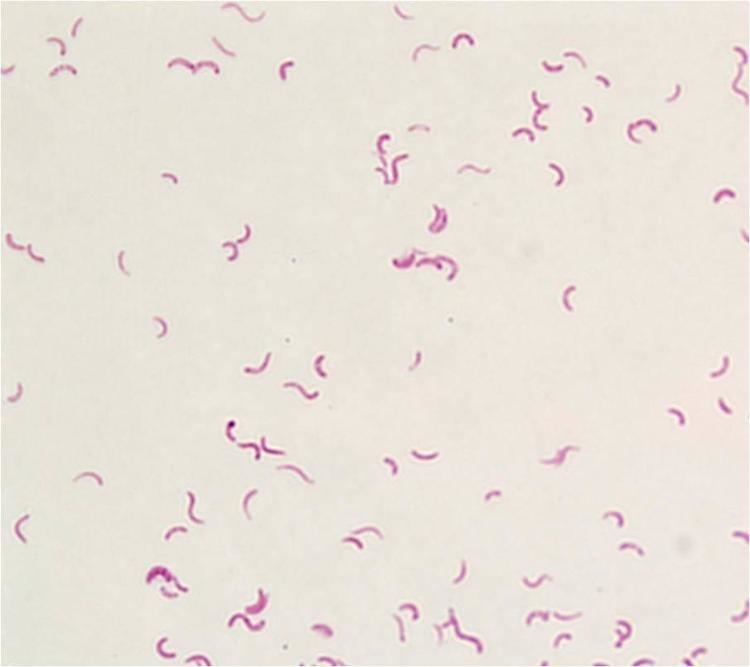
Rhodospirillum rubrum (R. rubrum) is a Gram-negative, pink-coloured Proteobacterium, with a size of 800 to 1000 nanometers.
It is a facultative anaerobe, it can therefore use alcoholic fermentation under low oxygen conditions or use aerobic respiration in aerobic conditions. Under aerobic growth photosynthesis is genetically suppressed and R. rubrum is then colorless. After the exhaustion of oxygen, R. rubrum immediately starts the production of photosynthesis apparatus including membrane proteins, bacteriochlorophylls and carotenoids, i.e. the bacterium becomes photosynthesis active. The repression mechanism for the photosynthesis is poorly understood. The photosynthesis of R. rubrum differs from that of plants as it possesses not chlorophyll a, but bacteriochlorophylls. While bacteriochlorophyll an absorbs light having a maximum wavelength of 800 to 925 nm, chlorophyll absorbs light having a maximum wavelength of 660 to 680 nm. R. rubrum is a spiral-shaped bacterium (spirillum, plural form: spirilla).
R. rubrum is also a nitrogen fixing bacterium, i.e., it can express and regulate nitrogenase, a protein complex that can catalyse the conversion of atmospheric dinitrogen into ammonia. Due to this important property, R. rubrum has been the test subject of many different groups, so as to understand the complex regulatory schemes required for this reaction to occur (, among others). It was in R. rubrum that, for the first time, post-translational regulation of nitrogenase was demonstrated. Nitrogenase is modified by an ADP-ribosylation in the arginine residue 101 (Arg101) in response to the so-called "switch-off" effectors - glutamine or ammonia - and darkness.
R. rubrum has several potential uses in biotechnology: