 | ||
RDE-1 (RNAi-DEfective 1) is a primary Argonaute protein required for RNA-mediated interference (RNAi) in Caenorhabditis elegans. The rde-1 gene locus was first characterized in C. elegans mutants resistant to RNAi, and is a member of a highly conserved Piwi gene family that includes plant, Drosophila, and vertebrate homologs.
Contents
Primary siRNA-argonaute complex
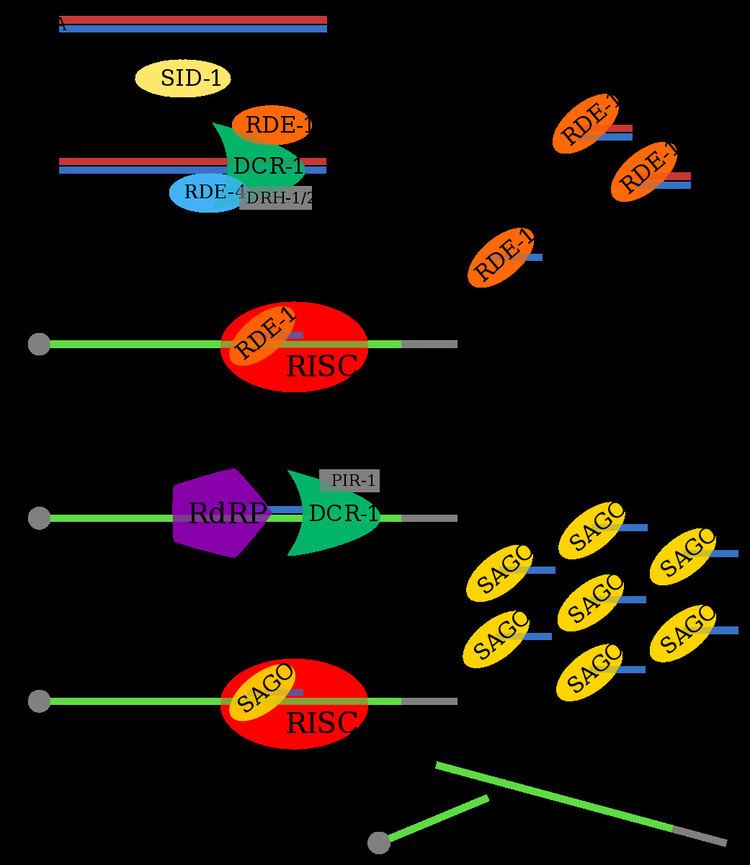
Upon uptake into the cell, exogenous trigger dsRNA is bound by RDE-4 and cleaved into 21-25nt primary siRNA by a Dicer complex that includes RDE-1. Primary siRNA binding to RDE-1 then promotes the formation of the RNA-induced silencing complex (RISC). Unlike in other Argonautes characterized in Drosophila and humans, the catalytic RNase H motif in RDE-1 has not been shown to exhibit Slicer activity of the target transcript. Instead, RNase H activity in RDE-1 is primarily facilitates siRNA maturation through cleavage of the passenger strand.
Secondary siRNA-argonaute complex
The primary Argonaute complex recruits an RNA-dependent RNA polymerase (RdRP) to generate secondary siRNAs, triggering an amplification response. Secondary siRNAs are bound by degenerate secondary Argonautes, which are then directly involved in target transcript degradation. However, RDE-1 shows no stable association with the more abundant secondary siRNAs.
Whereas rde-4 deficiency can be rescued by high concentrations of trigger dsRNA, and secondary Argonaute exhibit functional redundancy, there has been no evidence that RNA-mediated silencing can be reinstated in rde-1 deficient mutants. To this extent, RDE-1 appears to have a qualitiatively distinct activity in the exogenous RNAi pathway.
Structure
Canonical Argonaute proteins possess three primary domains forming a crescent-shaped base: the PAZ, MID, and PIWI domains. PAZ and MID orient and anchor the double-stranded siRNA by binding to the 3’ and 5’ termini, respectively, leaving the internal nucleotides accessible for base pairing. The PIWI domain folds into an RNase H-like structure, and contains the conserved catalytic triad “DDH” (two aspartate residues, one histidine residue). The crystal structure of RDE-1 has not been formally elucidated, but can be assumed to closely resemble its human homologs.
Passenger strand degradation
Mutation of any residues in the RNase H catalytic triad abolishes Slicer activity in human Argonaute protein Ago2, suggesting that the RNase H domain is directly responsible for target mRNA degradation. However, RDE-1 has not been implicated in mRNA-cleavage activity.
Instead, RDE-1 with mutations in the conserved DDH motif exhibit reduced passenger (sense) strand turnover, suggesting that RNase H activity serves to cleave the passenger strand, leaving the guide (antisense) strand accessible for base-pairing to target mRNA. Further, target silencing can be fully restored in DDH motif mutants by loading single-stranded siRNA, suggesting that a downstream component in the RNAi pathway is responsible for Slicer activity. Thus, RDE-1’s RNase H domain facilitates siRNA maturation but is not directly involved in cleaving target mRNA transcripts.
