 | ||
Prp24 (precursor RNA processing, gene 24) is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.
Contents
Biological Role
The process of spliceosome formation involves the U4 and U6 snRNPs associating and forming a di-snRNP in the cell nucleus. This di-snRNP then recruits another member (U5) to become a tri-snRNP. U6 must then dissociate from U4 to bond with U2 and become catalytically active. Once splicing has been done, U6 must dissociate from the spliceosome and bond back with U4 to restart the cycle.
Prp24 has been shown to promote the binding of U4 and U6 snRNPs. Removing Prp24 results in the accumulation of free U4 and U6, and the subsequent addition of Prp24 regenerates U4/U6 and reduces the amount of free U4 and U6. Naked U6 snRNA is very compact and has little room to form base pairs with other RNA. However, when U6 snRNP associates with proteins such as Prp24, the structure is much more open, thus facilitating the binding to U4. Prp24 is not present in the U6/U4 duplex itself, and it has been suggested that Prp24 must leave the complex in order for proper base pairs to be formed. It has also been suggested that Prp24 may play a role in destabilizing U4/U6 in order for U6 to pair bases with U2.
Structure
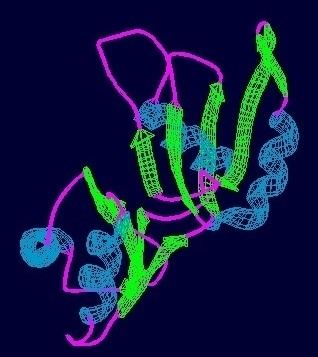
Prp24 has a molecular weight of 50 kDa and has been shown to contain four RNA recognition motifs (RRMs) and a conserved 12-amino acid sequence at the C-terminus. RRMs 1 and 2 have been shown to be important for high-affinity binding of U6, while RRMs 3 and 4 bind at lower affinity sites on U6. The first three RRMs interact extensively with each other and contain canonical folds that contain a four-stranded beta-sheet and two alpha-helices. The electropositive surface of RRMs 1 and 2 is a RNA annealing domain while the cleft between RRMs 1 and 2 including the beta-sheet face of RRM2 is a sequence-specific RNA binding site. The C-terminal motif is required for association with LSm proteins and contributes to substrate (U6) binding and not the catalytic rate of splicing.
Interactions
Prp24 interacts with the U6 snRNA via its RRMs. It has been shown through chemical modification testing that nucleotides 39–57 of U6 (40–43 in particular) are involved in binding Prp24.
The LSm proteins are in a consistent configuration on the U6 RNA. It has been proposed that the LSm proteins and Prp24 interact both physically and functionally and the C-terminal motif of Prp24 is important for this interaction. The binding of Prp24 to U6 is enhanced by the binding of Lsm proteins to U6, as is binding of U4 and U6. It was revealed by electron microscopy that Prp24 may interact with the LSm protein ring at LSm2.
Homologs
Prp24 has a human homolog, SART3. SART3 is a tumor rejection antigen (SART3 stands for "squamous cell carcinoma antigen recognized by T cells, gene 3). The RRMs 1 and 2 in yeast are similar to RRMs in human SART3. The C-terminal domain is also highly conserved from yeast to humans. This protein, like Prp24, interacts with the LSm proteins for the recycling of U6 into the U4/U6 snRNP. It has been proposed that SART3 target U6 to a Cajal body or a nuclear inclusion as the site of assembly of the U4/U6 snRNP. SART3 is located on chromosome 12, and a mutation is likely the cause of disseminated superficial actinic porokeratosis.
