Symbol PPARA Entrez 5465 OMIM 170998 | Alt. symbols PPAR HUGO 9232 RefSeq NM_001001928 | |
 | ||
In the field of molecular biology, the peroxisome proliferator-activated receptors (PPARs) are a group of nuclear receptor proteins that function as transcription factors regulating the expression of genes. PPARs play essential roles in the regulation of cellular differentiation, development, and metabolism (carbohydrate, lipid, protein), and tumorigenesis of higher organisms.
Contents
Nomenclature and tissue distribution
Three types of PPARs have been identified: alpha, gamma, and delta (beta):
History
PPARs were originally identified in Xenopus frogs as receptors that induce the proliferation of peroxisomes in cells. The first PPAR (PPARα) was discovered during the search of a molecular target for a group of agents then referred to as peroxisome proliferators, as they increased peroxisomal numbers in rodent liver tissue, apart from improving insulin sensitivity. These agents, pharmacologically related to the fibrates were discovered in the early 1980s. When it turned out that PPARs played a much more versatile role in biology, the agents were in turn termed PPAR ligands. The best-known PPAR ligands are the thiazolidinediones; see below for more details.
After PPARδ (delta) was identified in humans in 1992, it turned out to be closely related to the PPARβ (beta) previously described during the same year in other animals (Xenopus). The name PPARδ is generally used in the US, whereas the use of the PPARβ denomination has remained in Europe where this receptor was initially discovered in Xenopus.
Physiological function
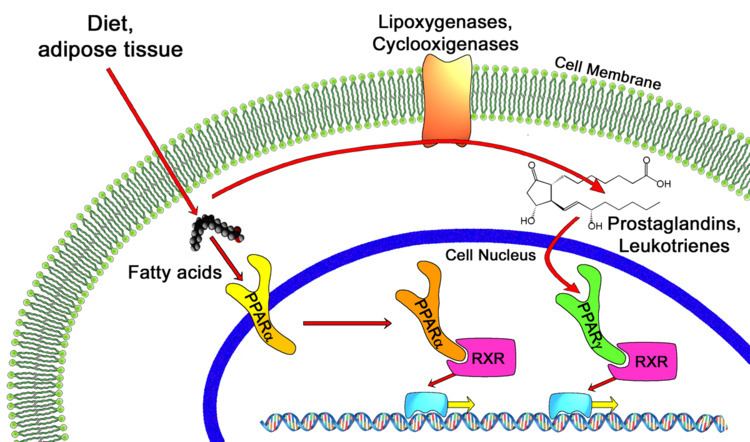
All PPARs heterodimerize with the retinoid X receptor (RXR) and bind to specific regions on the DNA of target genes. These DNA sequences are termed PPREs (peroxisome proliferator hormone response elements). The DNA consensus sequence is AGGTCANAGGTCA, with N being any nucleotide. In general, this sequence occurs in the promotor region of a gene, and, when the PPAR binds its ligand, transcription of target genes is increased or decreased, depending on the gene. The RXR also forms a heterodimer with a number of other receptors (e.g., vitamin D and thyroid hormone).
The function of PPARs is modified by the precise shape of their ligand-binding domain (see below) induced by ligand binding and by a number of coactivator and corepressor proteins, the presence of which can stimulate or inhibit receptor function, respectively.
Endogenous ligands for the PPARs include free fatty acids and eicosanoids. PPARγ is activated by PGJ2 (a prostaglandin) and certain members of the 5-HETE family of arachidonic acid metabolites including 5-oxo-15(S)-HETE and 5-oxo-ETE. In contrast, PPARα is activated by leukotriene B4. Certain members of the 15-Hydroxyicosatetraenoic acid family of arachidonic acid metabolites, including 15(S)-HETE, 15(R)-HETE, and 15-HpETE activate to varying degrees PPAR alpha, beta/delta, and gamma. PPARγ activation by agonist RS5444 may inhibit anaplastic thyroid cancer growth. See for a review and critique of the roles of PPAR gamma in cancer.
Genetics
The three main forms are transcribed from different genes:
Hereditary disorders of all PPARs have been described, generally leading to a loss in function and concomitant lipodystrophy, insulin resistance, and/or acanthosis nigricans. Of PPARγ, a gain-of-function mutation has been described and studied (Pro12Ala) which decreased the risk of insulin resistance; it is quite prevalent (allele frequency 0.03 - 0.12 in some populations). In contrast, pro115gln is associated with obesity. Some other polymorphisms have high incidence in populations with elevated body mass indexes.
Structure
Like other nuclear receptors, PPARs are modular in structure and contain the following functional domains:
The DBD contains two zinc finger motifs, which bind to specific sequences of DNA known as hormone response elements when the receptor is activated. The LBD has an extensive secondary structure consisting of 13 alpha helices and a beta sheet. Natural and synthetic ligands bind to the LBD, either activating or repressing the receptor.
Pharmacology and PPAR modulators
PPARα and PPARγ are the molecular targets of a number of marketed drugs. For instance the hypolipidemic fibrates activate PPARα, and the anti diabetic thiazolidinediones activate PPARγ. The synthetic chemical perfluorooctanoic acid activates PPARα while the synthetic perfluorononanoic acid activates both PPARα and PPARγ. Berberine activates PPARγ, as well as other natural compounds from different chemical classes.
