Entrez 51236 | Ensembl ENSG00000235173 | |
 | ||
Aliases HGH1, BRP16, BRP16L, C8orf30A, C8orf30B, FAM203A, FAM203B, HGH1 homolog External IDs MGI: 1930628 HomoloGene: 48742 GeneCards: HGH1 | ||
Family with Sequence Similarity 203, Member B (FAM203B) is a protein encoded by the FAM203B gene (8q24.3) in humans. While FAM203B is only found in humans and possibly non-human primates, its paralog, FAM203A, is highly conserved. The FAM203B protein contains two conserved domains of unknown function, DUF383 and DUF384, and no transmembrane domains. This protein has no known function yet, although the homolog of FAM203A in Caenorhabditis elegans (Y54H5A.2) is thought to help regulate the actin cytoskeleton.
Contents
Gene
FAM203B is located on the positive DNA strand of the long arm of chromosome 8 at locus 24.3 (8q24.3) from 76,368,898 - 76,371,411 in the human genome. The gene product contains 2,402 bp of mRNA with 6 predicted exons in the human gene. There are no known isoforms.
Gene Neighborhood
The pseudogene TSSK5P2 is located on the negative strand opposite FAM203B (145,440,975 - 145,443,775), while LOC377711 is located immediately downstream on the positive strand (145,448,755 - 145,485,896). FAM203A, MROH1, and SCXB are located upstream of FAM203B.
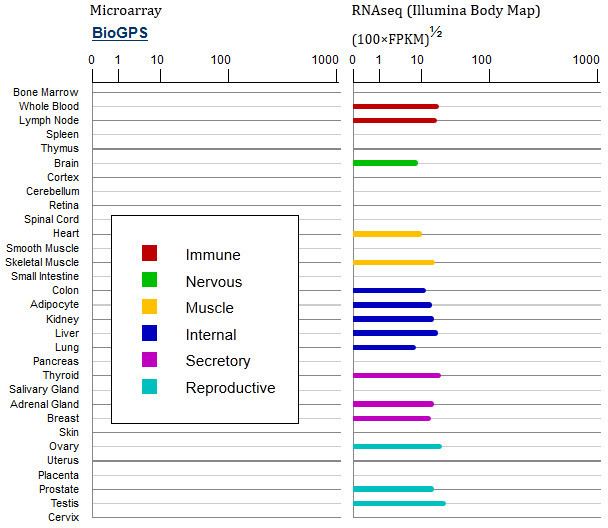
Gene Expression
Expression Profile: mRNA expression has been localized in many tissue types (immune, nervous, muscle, internal, secretory, and reproductive) in similar quantities and may therefore be ubiquitous.
Promoter: The predicted promoter region of FAM203B is located between 145,437,380 and 145,438,015 on Chromosome 8 and has a length of 636 bp.
Protein
The function of FAM203B is not currently understood. The FAM203B protein has 390 amino acids, a molecular weight of 42.1 kdal, and an isoelectric point of 4.56.
Structure
FAM203B contains two domains of unknown function: DUF383 (residues 110-288) and DUF384 (residues 292-349). The protein is alanine-, proline-, and leucine-rich, but poor in serine, asparagine, threonine, isoleucine, lysine, and phenylalanine. The following internal repeats can be found in the primary sequence: LPFL (26-29, 245-248), ELAP (70-73), GRAL (54-57, 111-114), and LAADPGL (88-94, 99-105). There are no positive, negative, mixed charge, or hydrophobic clusters; no transmembrane domains; and no clusters of amino acid multiplets. The secondary structure prediction generated by the Phyre 2.0 bioinformatic server shows only α-helices, almost all of which have high confidence values. The overall confidence value of the model is 99.5%.
Post-Translational Modifications
There are at least six predicted phosphorylation sites in FAM203B: S17, S153, Y167, T223, S259, and S320. The FAM203B protein is also predicted to locate to the cytoplasm.
Protein Interactions
There are many possible transcription factor binding sites in the FAM203B promoter. Below is a table of the best possibilities, which have high confidence values, evolutionary conservation, and/or multiple possible binding sites in the promoter.
Table of Possible Transcription Factor Binding Sites in Predicted FAM203B Promoter:
There are several other proteins that may interact directly with the FAM203B protein including C1orf112, HEATR3, MRTO4, BYSL, GINS1, DKC1, TXNDC12, PWP2, IMP4, and NIP7.
FAM203A: Paralog
FAM203A is 99% identical to FAM203B with only one amino acid difference (E264Q) due to a point mutation (G857C). This indicates that the duplication event that produced FAM203B 242,266 bp downstream from FAM203A occurred very recently in evolutionary history. The FAM203A protein is highly conserved and has orthologs in primates, rodents, ungulates, marsupials, amphibians, fish, fungi, plants, and at least one monotreme, one reptile, and one hemichordate.
Orthologs and Homologs
Table of FAM203B Paralog and Homologs:
There is one ortholog of FAM203B, brain protein 16-like (BRP16L) in Macaca mulatta, although no other primates appear to have orthologous proteins. There are two possible explanations for this anomaly: (1) DNA of other primates has not been sequenced thoroughly in the genomic region of the FAM203B ortholog, or (2) FAM203B is the result of a gene duplication event unique to humans, meaning that BRP16L in M. mulatta resulted from an earlier duplication event unique to that species. The second explanation is supported by the following evidence:
- Like M. mulatta, Danio rerio has both a FAM203A gene and a BRP16L gene. The large amount of time since the divergence of the M. mulatta and D. rerio lineages suggests that these BRP16L genes are the result of separate duplication events.
- The BRP16L protein in D. rerio has a significant 3’ truncation compared to the M. mulatta protein, further supporting the hypothesis that these proteins evolved separately.
- If the BRP16L genes in "M mulatta" and "D. rerio" are the result of separate duplication events, then it is also possible that FAM203B and BRP16L in "M. mulatta" are the result of separate duplication events.
- BRP16 (brain protein 16) is an alias of FAM203A, and BRP16L (brain protein 16-like) is an alias of FAM203B. A gene named BRP16L simply means that the gene is related to FAM203A but not necessarily to FAM203B.
- FAM203A and FAM203B are located in the telomeric region of chromosome 8, an area of chromosomes that frequently experiences recombination events.
However, because FAM203A and FAM203B are so similar, it is difficult to determine whether proteins are orthologs or just simply homologs.
Phylogeny
The phylogenetic tree of FAM203B and its homologs matches with the overall divergence of the respective lineages.
Conserved Domains, Motifs, and Residues
- ARM (armadillo/beta-catenin-like repeats-containing): Found in two homologs (FAM203A in Danio rerio and At1g14300 in Arabidopsis thaliana) and overlaps slightly with the beginning of the DUF383 domain. Related to the HEAT domain, consists of a 40-amino-acid tandemly repeated sequence motif, and is thought to mediate protein-protein interactions. Several eukaryotic genes contain ARM domains including armadillo in Drosophila melanogaster, beta-catenin, plakoglobin, and adenomatous polyposis coli in mammals.
- DUF383: Domain of unknown function 383
- DUF384: Domain of unknown function 383
Every ortholog and homolog of FAM203B has a DUF383 domain and a DUF384 domain (except Anolis carolinensis, which is missing DUF384 due to a large 3' truncation). There is significant variation among mammals, marsupials, and monotremes as to where the DUF383 domain begins, whereas this variation is smaller in reptiles, amphibians, fish, invertebrates, plants, and fungi. Additionally, the DUF383 domain ends at the same location for all homologs, while the DUF384 domain starts and ends at roughly the same location in all homologs. There is high homology in the DUF384 domain (292..349) and in the DUF383 domain (154..288), and several amino acids are completely conserved in vertebrates, invertebrates, plants, and fungi, which include Arg190, Gly219, Asn226, Lys273, and Lys338. Other highly conserved amino acids include Asn87, Lys88, Arg216, and Phe229.
