Entrez 221786 | Ensembl ENSG00000221909 | |
 | ||
Aliases FAM200A, C7orf38, family with sequence similarity 200 member A External IDs HomoloGene: 89159 GeneCards: FAM200A | ||
C7orf38 is a gene located on chromosome 7 in the human genome. The gene is expressed in nearly all tissue types at very low levels. Evolutionarily, it can be found throughout the kingdom animalia. While the function of the protein is not fully understood by the scientific community, bioinformatic tools have shown that the protein bares much similarity to zinc finger or transposase proteins. Many of its orthologs, paralogs, and neighboring genes have been shown to possess zinc finger domains. The protein contains a hAT dimerization domain nears its C-terminus. This domain is highly conserved in transposase enzymes.
Contents
Gene
C7orf38 is located on chromosome 7 at q22.1. Its genomic sequence contains 5,612 bp. The predominant transcript contains two exons and is 2,507 bp in length. The translated protein contains 573 amino acids.
Protein composition
The 573 amino acid protein has a molecular weight of 66,280.05. The isoelectric point was found to occur at a pH of 5.775, about 1.6 pH lower than that of the average human pH. Two deviations from prototypical human proteins are evident. The protein contains a less than expected number of glycine residues, and is rich in leucine residues. There are not sections of strong hydrophobicity or hydrophilicity. Thus, it is not predicted to be a transmembrane protein.
Gene neighborhood
The four genes in closest proximity to C7orf38 on chromosome 7 exhibit similar function, many of which are transcription factors.
Paralogs
Eight paralogs are found in the human proteome. Similar to the neighboring genes, many of the paralogs function as zinc fingers, or transcription factors.
Orthologs
Orthologs to C7orf38 can be traced back evolutionarily through plants. The following is not an extensive list of orthologs. It is intended to provide an evolutionary overview of the conservation of C7orf38.
Protein
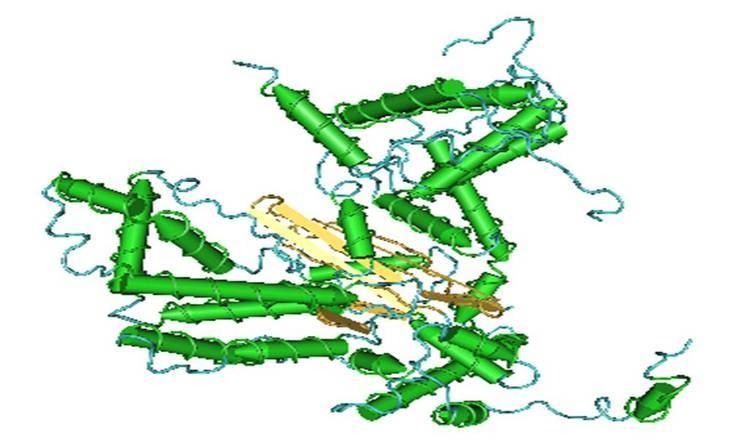
CBLast was used to determine a structurally related protein with experimentally determined structure. The protein Hermes DNA transposase, of the Hermes DBD superfamily, was shown to be structurally similar (Evalue: 1E-6).
The hAT dimerization domain is found at the C-terminus of transposase elements belonging to the Activator superfamily (hAT element superfamily). The isolated dimerization domain forms extremely stable dimers in vitro.
mRNA
The MFOLD program available at Rensselaer BioInformatics Server was used to predict secondary structure of the mature mRNA sequence. The primary sequence of the mRNA secondary structures displayed high levels of conservation in orthologs, suggesting structural importance.
Tissue distribution
The gene appears to be expressed in most tissue types. Very low levels of expression were observed through est profiles, and no deviation was observed between health or developmental states.
