Symbol DPS PDB 1QGH | Entrez 1129452 UniProt P80725 | |
 | ||
DNA-binding proteins from starved cells (DPS) are proteins that belong to the ferritin superfamily and are characterized by strong similarities but also distinctive differences with respect to "canonical" ferritins.
Contents
DPS proteins are part of a complex bacterial defence system that protects DNA against oxidative damage and are distributed widely in the bacterial kingdom.
Description
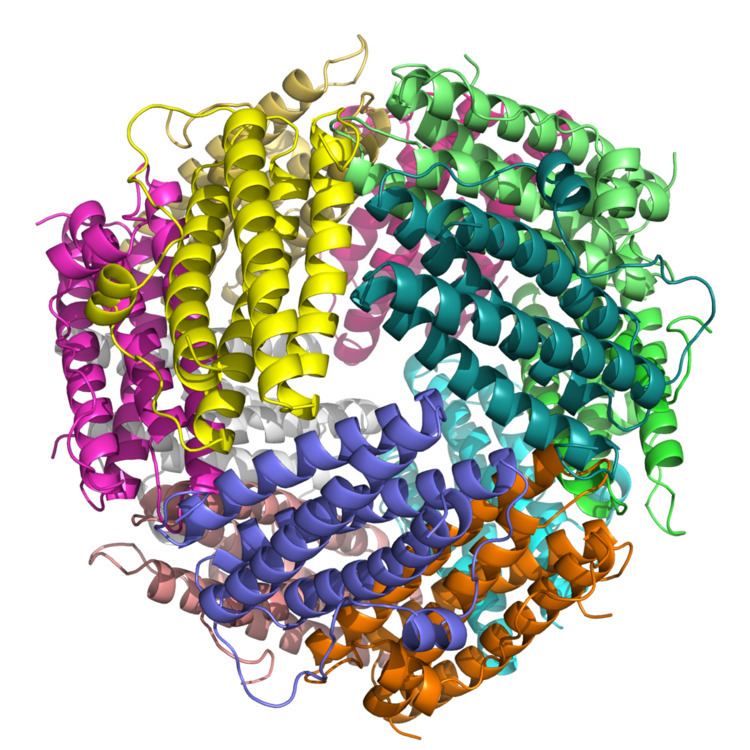
DPS are highly symmetrical dodecameric proteins of 200 kDa characterized from a shell-like structure of 2:3 tetrahedral symmetry assembled from identical subunits with an external diameter of ~ 9 nm and a central cavity of ~ 4.5 nm in diameter. Dps proteins belong to the ferritin superfamily and the DNA protection is afforded by means of a double mechanism:
The first was discovered in Escherichia coli Dps in 1992 and has given the name to the protein family; during stationary phase, Dps binds the chromosome non-specifically, forming a highly ordered and stable dps-DNA co-crystal within which chromosomal DNA is condensed and protected from diverse damages. The lysine-rich N-terminus is required for self-aggregation as well as for Dps-driven DNA condensation.
The second mode of protection is due to the ability of Dps proteins to bind and oxidize Fe(II) at the characteristic, highly conserved intersubunit ferroxidase center.
The dinuclear ferroxidase centers are located at the interfaces between subunits related by 2-fold symmetry axes. Fe(II) is sequestered and stored in the form of Fe(III) oxyhydroxide mineral, which can be released after reduction. In the mineral iron core up to 500 Fe(III) can be deposited. One hydrogen peroxide oxidizes two Fe2+ ions, which prevents hydroxyl radical production by the Fenton reaction (reaction I):
2 Fe2+ + H2O2 + 2 H+ = 2 Fe3+ + 2 H2O
Dps also protects the cell from UV and gamma ray irradiation, iron and copper toxicity, thermal stress and acid and base shocks. Also shows a weak catalase activity.
DNA condensation
Dps dodecamers can condense DNA in vitro through a cooperative binding mechanism. Deletion of portions of the N-terminus or mutation of key lysine residues in the N-terminus can impair or eliminate the condensation activity of Dps. Single molecule studies have shown that Dps-DNA complexes can get trapped in long-lived metastable states that exhibit hysteresis. Because of this, the extent of DNA condensation by Dps can depend not only on the current buffer conditions but also on the conditions in the past. A modified Ising model can be used to explain this binding behavior.
Expression
In Escherichia coli Dps protein is Induced by rpoS and IHF in the early stationary phase. Dps is also Induced by oxyR in response to oxidative stress during exponential phase. ClpXP probably directly regulate proteolysis of dps during exponential phase. ClpAP seems to play an indirect role in maintaining ongoing dps synthesis during stationary phase
Applications
Cavities formed by Dps and ferritin proteins have been successfully used as the reaction chamber for the fabrication of metal nanoparticles (NPs). Protein shells served as a template to restrain particle growth and as a coating to prevent coagulation/aggregation between NPs. Using various sizes of protein shells, various sizes of NPs can be easily synthesized for chemical, physical and bio-medical applications.
