Entrez 4512 | Ensembl ENSG00000198804 | |
 | ||
Aliases COX1, mitochondrially encoded cytochrome c oxidase I, COI, MTCO1, Main subunit of cytochrome c oxidase, CO I, cytochrome c oxidase subunit I External IDs MGI: 102504 HomoloGene: 5016 GeneCards: COX1 | ||
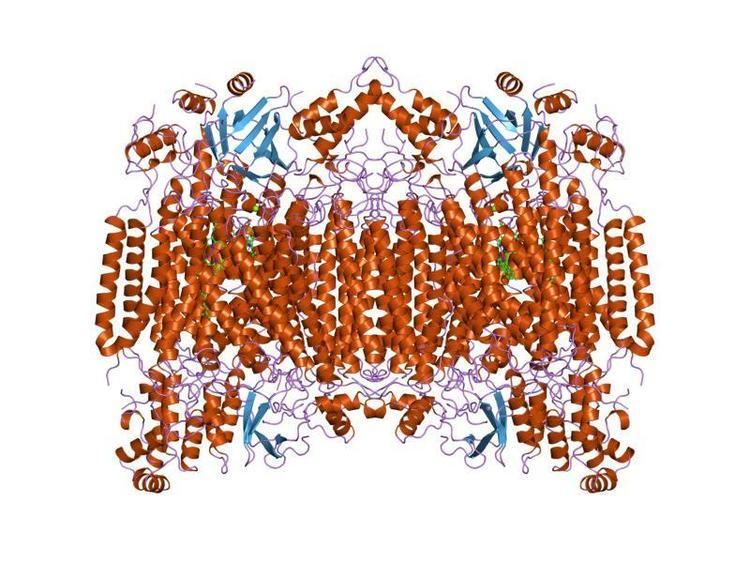
Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that in humans is encoded by the MT-CO1 gene. In other eukaryotes, the gene is called COX1, CO1, or COI. Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex.
Contents
Function
Cytochrome c oxidase subunit I (CO1 or MT-CO1) is one of three mitochondrial DNA (mtDNA) encoded subunits (MT-CO1, MT-CO2, MT-CO3) of respiratory complex IV. Complex IV is the third and final enzyme of the electron transport chain of mitochondrial oxidative phosphorylation.
Cytochrome c oxidase (EC 1.9.3.1) is a key enzyme in aerobic metabolism. Proton pumping heme-copper oxidases represent the terminal, energy-transfer enzymes of respiratory chains in prokaryotes and eukaryotes. The CuB-heme a3 (or heme o) binuclear centre, associated with the largest subunit I of cytochrome c and ubiquinol oxidases (EC 1.10.3.10), is directly involved in the coupling between dioxygen reduction and proton pumping. Some terminal oxidases generate a transmembrane proton gradient across the plasma membrane (prokaryotes) or the mitochondrial inner membrane (eukaryotes).
The enzyme complex consists of 3-4 subunits (prokaryotes) up to 13 polypeptides (mammals) of which only the catalytic subunit (equivalent to mammalian subunit I (COI)) is found in all heme-copper respiratory oxidases. The presence of a bimetallic centre (formed by a high-spin heme and copper B) as well as a low-spin heme, both ligated to six conserved histidine residues near the outer side of four transmembrane spans within COI is common to all family members. In contrast to eukaryotes the respiratory chain of prokaryotes is branched to multiple terminal oxidases. The enzyme complexes vary in heme and copper composition, substrate type and substrate affinity. The different respiratory oxidases allow the cells to customize their respiratory systems according to a variety of environmental growth conditions.
It has been shown that eubacterial quinol oxidase was derived from cytochrome c oxidase in Gram-positive bacteria and that archaebacterial quinol oxidase has an independent origin. A considerable amount of evidence suggests that proteobacteria (Purple bacteria) acquired quinol oxidase through a lateral gene transfer from Gram-positive bacteria.
A related nitric-oxide reductase (EC 1.7.99.7) exists in denitrifying species of archaea and eubacteria and is a heterodimer of cytochromes b and c. Phenazine methosulphate can act as acceptor. It has been suggested that cytochrome c oxidase catalytic subunits evolved from ancient nitric oxide reductases that could reduce both nitrogen and oxygen.
Subfamilies
Application
It is a gene that is often used as a DNA barcode to identify animal species. MT-CO1 gene sequence is suitable for this role because its mutation rate is often fast enough to distinguish closely related species and also because its sequence is conserved among conspecifics. Contrary to the primary objection raised by skeptics that MT-CO1 sequence differences are too small to be detected between closely related species, more than 2% sequence divergence is typically detected between such organisms, suggesting that the barcode is effective. In most if not all seed plants, however, the rate of evolution of cox1 is very slow.
MT-COI (CCOI) in colonic crypts
CCOI is a synonym for MT-COI
CCOI protein is usually expressed at a high level in the cytoplasm of colonic crypts of the human large intestine (colon). However, CCOI is frequently lost in colonic crypts with age in humans and is also often absent in field defects that give rise to colon cancers as well as in portions of colon cancers.
The epithelial inner surface of the colon is punctuated by invaginations, the colonic crypts. The colon crypts are shaped like microscopic thick walled test tubes with a central hole down the length of the tube (the crypt lumen). Four tissue sections are shown in the image in this section, two cut across the long axes of the crypts and two cut parallel to the long axes.
Most of the human colonic crypts in the images have high expression of the brown-orange stained CCOI. However, in some of the colonic crypts all of the cells lack CCOI and appear mostly white, with their main color being the blue-gray staining of the nuclei at the outer walls of the crypts. Greaves et al. showed that deficiencies of CCOI in colonic crypts are due to mutations in the CCOI gene. As seen in panel B, a portion of the stem cells of three crypts appear to have a mutation in CCOI, so that 40% to 50% of the cells arising from those stem cells form a white segment in the cross-cut area.
In humans, the percent of colonic crypts deficient for CCOI is less than 1% before age 40, but then increases linearly with age. On average, the percent of colonic crypts deficient for CCOI reaches 18% in women and 23% in men by 80–84 years of age. Colonic tumors often arise in a field of crypts containing a large cluster (as many as 410) of CCOI-deficient crypts. In colonic cancers, up to 80% of tumor cells can be deficient in CCOI.
As seen in panels C and D, crypts are about 75 to about 110 cells long. The average crypt circumference is 23 cells. Based on these measurements, crypts have between 1725 and 2530 cells. Another report gave a range of 1500 to 4900 cells per colonic crypt.
The occurrence of frequent crypts with almost complete loss of CCOI in their 1700 to 5,000 cells suggests a process of natural selection. However, it has also been shown that a deficiency throughout a particular crypt due to an initial mitochondrial DNA mutation may occasionally occur through a stochastic process. Nevertheless, the frequent occurrence of CCOI deficiency in many crypts within a colon epithelium indicates that absence of CCOI likely provides a selective advantage.
CCOI is coded for by the mitochondrial chromosome. There are multiple copies of the chromosome in most mitochondria, usually between 2 and 6 per mitochondrion. If a mutation occurs in CCOI in one chromosome of a mitochondrion, there may be random segregation of the chromosomes during mitochondrial fission to generate new mitochondria. This can give rise to a mitochondrion with primarily or solely CCOI-mutated chromosomes.
A mitochondrion with largely CCOI-mutated chromosomes would need to have a positive selection bias in order to frequently become the main type of mitochondrion in a cell (a cell with CCOI-deficient homoplasmy). There are about 100 to 700 mitochondria per cell, depending on cell type. Furthermore, there is fairly rapid turnover of mitochondria, so that a mitochondrion with CCOI-mutated chromosomes and a positive selection bias could shortly become the major type of mitochondrion in a cell. The average half-life of mitochondria in rats, depending on cell type, is between 9 and 24 days, and in mice is about 2 days. In humans it is likely that the half life of mitochondria is also a matter of days to weeks.
A stem cell at the base of a colonic crypt that was largely CCOI-deficient may compete with the other 4 or 5 stem cells to take over the stem cell niche. If this occurs, then the colonic crypt would be deficient in CCOI in all 1700 to 5,000 cells, as is indicated for some crypts in panels A, B and D of the image.
Crypts of the colon can reproduce by fission, as seen in panel C, where a crypt is fissioning to form two crypts, and in panel B where at least one crypt appears to be fissioning. Most crypts deficient in CCOI are in clusters of crypts (clones of crypts) with two or more CCOI-deficient crypts adjacent to each other (see panel D). This illustrates that clones of deficient crypts often arise, and thus that there is likely a positive selective bias that has allowed them to spread in the human colonic epithelium.
It is not clear why a deficiency of CCOI should have a positive selective bias. One suggestion is that deficiency of CCOI in a mitochondrion leads to lower reactive oxygen production (and less oxidative damage) and this provides a selective advantage in competition with other mitochondria within the same cell to generate homoplasmy for CCOI-deficiency. Another suggestion was that cells with a deficiency in cytochrome c oxidase are apoptosis resistant, and thus more likely to survive. The linkage of CCOI to apoptosis arises because active cytochrome c oxidase oxidizes cytochrome c, which then activates pro-caspase 9, leading to apoptosis. These two factors may contribute to the frequent occurrence of CCOI-deficient colonic crypts with age or during carcinogenesis in the human colon.
