 | ||
Computational anatomy is a discipline within medical imaging focusing on the study of anatomical shape and form at the visible or gross anatomical scale of morphology. It involves the development and application of computational, mathematical and data-analytical methods for modeling and simulation of biological structures.
Contents
- Genesis
- History
- The deformable template orbit model of computational anatomy
- Shapes and forms
- Groups and group actions
- Lagrangian and Eulerian flows for generating diffeomorphisms
- The diffeomorphism group of computational anatomy
- Diffeomorphometry The metric space of shapes and forms
- The right invariant metric on diffeomorphisms
- The metric on shapes and forms
- The action integral for Hamiltons principle on diffeomorphic flows
- Diffeomorphic or Eulerian shape momentum
- The EulerLagrange equation on shape momentum for geodesics on the group of diffeomorphisms
- Diffeomorphic shape momentum a classical vector function
- Riemannian exponential geodesic positioning and Riemannian logarithm geodesic coordinates
- Hamiltonian formulation of computational anatomy
- Stationarity of the Hamiltonian and kinetic energy along EulerLagrange
- The metric on geodesic flows of landmarks surfaces and volumes within the orbit
- Landmark or pointset geodesics
- Surface geodesics
- Volume geodesics
- Conservation laws on diffeomorphic shape momentum for computational anatomy
- Geodesic interpolation of information between coordinate systems via variational problems
- Matching based on minimizing kinetic energy action with endpoint condition
- Matching based on geodesic shooting
- Dense image matching in computational anatomy
- LDDMM dense image matching
- Hamiltonian LDDMM in the reduced advected state
- Diffusion tensor image matching in computational anatomy
- High Angular Resolution Diffusion Image HARDI matching in computational anatomy
- Metamorphosis
- Matching landmarks curves surfaces
- Landmark or point matching with correspondence
- Measure matching unregistered landmarks
- Curve matching
- Surface matching
- Growth and atrophy from longitudinal time series
- The random orbit model of computational anatomy
- The Bayesian model of computational anatomy
- Statistical shape theory in computational anatomy
- Template estimation from populations
- Software for diffeomorphic mapping
- Cloud software
- References
The field is broadly defined and includes foundations in anatomy, applied mathematics and pure mathematics, machine learning, computational mechanics, computational science, medical imaging, neuroscience, physics, probability, and statistics; it also has strong connections with fluid mechanics and geometric mechanics. Additionally, it complements newer, interdisciplinary fields like bioinformatics and neuroinformatics in the sense that its interpretation uses metadata derived from the original sensor imaging modalities (of which Magnetic Resonance Imaging is one example). It focuses on the anatomical structures being imaged, rather than the medical imaging devices. It is similar in spirit to the history of Computational linguistics, a discipline that focuses on the linguistic structures rather than the sensor acting as the transmission and communication medium(s).
In computational anatomy, the diffeomorphism group is used to study different coordinate systems via coordinate transformations as generated via the Lagrangian and Eulerian velocities of flow in
Genesis
At Computational anatomy's heart is the comparison of shape by recognizing in one shape the other. This connects it to D'Arcy Wentworth Thompson's developments On Growth and Form which has led to scientific explanations of morphogenesis, the process by which patterns are formed in Biology. Albrecht Durer's Four Books on Human Proportion were arguably the earliest works on Computational anatomy. The efforts of Noam Chomsky in his pioneering of Computational Linguistics inspired the original formulation of Computational anatomy as a generative model of shape and form from exemplars acted upon via transformations.
Due to the availability of dense 3D measurements via technologies such as magnetic resonance imaging (MRI), Computational anatomy has emerged as a subfield of medical imaging and bioengineering for extracting anatomical coordinate systems at the morphome scale in 3D. The spirit of this discipline shares strong overlap with areas such as computer vision and kinematics of rigid bodies, where objects are studied by analysing the groups responsible for the movement in question. Computational anatomy departs from computer vision with its focus on rigid motions, as the infinite-dimensional diffeomorphism group is central to the analysis of Biological shapes. It is a branch of the image analysis and pattern theory school at Brown University pioneered by Ulf Grenander. In Grenander's general Metric Pattern Theory, making spaces of patterns into a metric space is one of the fundamental operations since being able to cluster and recognize anatomical configurations often requires a metric of close and far between shapes. The diffeomorphometry metric of Computational anatomy measures how far two diffeomorphic changes of coordinates are from each other, which in turn induces a metric on the shapes and images indexed to them. The models of metric pattern theory, in particular group action on the orbit of shapes and forms is a central tool to the formal definitions in Computational anatomy.
History
Computational anatomy is the study of shape and form at the morphome or gross anatomy millimeter, or morphology scale, focusing on the study of sub-manifolds of
The first formalization of Computational Anatomy as an orbit of exemplar templates under diffeomorphism group action was in the original lecture given by Grenander and Miller with that title in May 1997 at the 50th Anniversary of the Division of Applied Mathematics at Brown University, and subsequent publication. This was the basis for the strong departure from much of the previous work on advanced methods for spatial normalization and image registration which were historically built on notions of addition and basis expansion. The structure preserving transformations central to the modern field of Computational Anatomy, homeomorphisms and diffeomorphisms carry smooth submanifolds smoothly. They are generated via Lagrangian and Eulerian flows which satisfy a law of composition of functions forming the group property, but are not additive.
The original model of Computational anatomy was as the triple,
The Lagrangian and Hamiltonian formulations of the equations of motion of Computational Anatomy took off post 1997 with several pivotal meetings including the 1997 Luminy meeting organized by the Azencott school at Ecole-Normale Cachan on the "Mathematics of Shape Recognition" and the 1998 Trimestre at Institute Henri Poincaré organized by David Mumford "Questions Mathématiques en Traitement du Signal et de l'Image" which catalyzed the Hopkins-Brown-ENS Cachan groups and subsequent developments and connections of Computational anatomy to developments in global analysis.
The developments in Computational Anatomy included the establishment of the Sobelev smoothness conditions on the diffeomorphometry metric to insure existence of solutions of variational problems in the space of diffeomorphisms, the derivation of the Euler-Lagrange equations characterizing geodesics through the group and associated conservation laws, the demonstration of the metric properties of the right invariant metric, the demonstration that the Euler-Lagrange equations have a well-posed initial value problem with unique solutions for all time, and with the first results on sectional curvatures for the diffeomorphometry metric in landmarked spaces. Following the Los Alamos meeting in 2002, Joshi's original large deformation singular Landmark solutions in Computational anatomy were connected to peaked Solitons or Peakons as solutions for the Camassa-Holm equation. Subsequently, connections were made between Computational anatomy's Euler-Lagrange equations for momentum densities for the right-invariant metric satisfying Sobolev smoothness to Vladimir Arnold's characterization of the Euler equation for incompressible flows as describing geodesics in the group of volume preserving diffeomorphisms. The first algorithms, generally termed LDDMM for large deformation diffeomorphic mapping for computing connections between landmarks in volumes and spherical manifolds, curves, currents and surfaces, volumes, tensors, varifolds, and time-series have followed.
These contributions of Computational anatomy to the global analysis associated to the infinite dimensional manifolds of subgroups of the diffeomorphism group is far from trivial. The original idea of doing differential geometry, curvature and geodesics on infinite dimensional manifolds goes back to Bernhard Riemann's Habilitation (Ueber die Hypothesen, welche der Geometrie zu Grunde liegen); the key modern book laying the foundations of such ideas in global analysis are from Michor.
The applications within Medical Imaging of Computational Anatomy continued to flourish after two organized meetings at the Institute for Pure and Applied Mathematics conferences at University of California, Los Angeles. Computational anatomy has been useful in creating accurate models of the atrophy of the human brain at the morphome scale, as well as Cardiac templates, as well as in modeling biological systems. Since the late 1990s, computational anatomy has become an important part of developing emerging technologies for the field of medical imaging. Digital atlases are a fundamental part of modern Medical-school education and in neuroimaging research at the morphome scale. Atlas based methods and virtual textbooks which accommodate variations as in deformable templates are at the center of many neuro-image analysis platforms including Freesurfer, FSL, MRIStudio, SPM. Diffeomorphic registration, introduced in the 90's, is now an important player with existing codes bases organized around ANTS, DARTEL, DEMONS, LDDMM, StationaryLDDMM, FastLDDMM, are examples of actively used computational codes for constructing correspondences between coordinate systems based on sparse features and dense images. Voxel-based morphometry(VBM) is an important technology built on many of these principles.
The deformable template orbit model of computational anatomy
The model of human anatomy is a deformable template, an orbit of exemplars under group action. Deformable template models have been central to Grenander's Metric Pattern theory, accounting for typicality via templates, and accounting for variability via transformation of the template. An orbit under group action as the representation of the deformable template is a classic formulation from differential geometry. The space of shapes are denoted
The orbit
The orbit model of computational anatomy is an abstract algebra - to be compared to linear algebra- since the groups act nonlinearly on the shapes. This is a generalization of the classical models of linear algebra, in which the set of finite dimensional
Shapes and forms
The central objects are shapes or forms in Computational anatomy, one set of examples being the 0,1,2,3-dimensional submanifolds of
The 0-dimensional manifolds are landmarks or fiducial points; 1-dimensional manifolds are curves such as sulcul and gyral curves in the brain; 2-dimensional manifolds correspond to boundaries of substructures in anatomy such as the subcortical structures of the midbrain or the gyral surface of the neocortex; subvolumes correspond to subregions of the human body, the heart, the thalamus, the kidney.
The landmarks
Groups and group actions
Groups and group actions are familiar to the Engineering community with the universal popularization and standardization of linear algebra as a basic model for analyzing signals and systems in mechanical engineering, electrical engineering and applied mathematics. In linear algebra the matrix groups (matrices with inverses) are the central structure, with group action defined by the usual definition of
The central group in Computational anatomy defined on volumes in
Most popular are scalar images,
For sub-manifolds
Several group actions in computational anatomy have been defined.
Lagrangian and Eulerian flows for generating diffeomorphisms
For the study of rigid body kinematics, the low-dimensional matrix Lie groups have been the central focus. The matrix groups are low-dimensional mappings, which are diffeomorphisms that provide one-to-one correspondences between coordinate systems, with a smooth inverse. The matrix group of rotations and scales can be generated via a closed form finite-dimensional matrices which are solution of simple ordinary differential equations with solutions given by the matrix exponential.
For the study of deformable shape in Computational anatomy, a more general diffeomorphism group has been the group of choice, which is the infinite dimensional analogue. The high-dimensional differeomorphism groups used in Computational Anatomy are generated via smooth flows
with
The inverse
The diffeomorphism group of computational anatomy
The group of diffeomorphisms is very big. To ensure smooth flows of diffeomorphisms avoiding shock-like solutions for the inverse, the vector fields must be at least 1-time continuously differentiable in space. For diffeomorphisms on
The diffeomorphism group are flows with vector fields absolutely integrable in Sobolev norm:
where
Diffeomorphometry: The metric space of shapes and forms
The study of metrics on groups of diffeomorphisms and the study of metrics between manifolds and surfaces has been an area of significant investigation. In Computational anatomy, the diffeomorphometry metric measures how close and far two shapes or images are from each other. Informally, the metric length is the shortest length of the flow which carries one coordinate system into the other.
Oftentimes, the familiar Euclidean metric is not directly applicable because the patterns of shapes and images don't form a vector space. In the Riemannian orbit model of Computational anatomy, diffeomorphisms acting on the forms
The right-invariant metric on diffeomorphisms
Define the distance on the group of diffeomorphisms
this is the right-invariant metric of diffeomorphometry, invariant to reparameterization of space since for all
The metric on shapes and forms
The distance on shapes and forms,
the images are denoted with the orbit as
The action integral for Hamilton's principle on diffeomorphic flows
In classical mechanics the evolution of physical systems is described by solutions to the Euler–Lagrange equations associated to the Least-action principle of Hamilton. This is a standard way, for example of obtaining Newton's laws of motion of free particles. More generally, the Euler-Lagrange equations can be derived for systems of generalized coordinates. The Euler-Lagrange equation in Computational anatomy describes the geodesic shortest path flows between coordinate systems of the diffeomorphism metric. In Computational anatomy the generalized coordinates are the flow of the diffeomorphism and its Lagrangian velocity
the Lagrangian is given by the kinetic energy:
Diffeomorphic or Eulerian shape momentum
In computational anatomy,
The Euler–Lagrange equation on shape momentum for geodesics on the group of diffeomorphisms
Classical calculation of the Euler-Lagrange equation from Hamilton's principle requires the perturbation of the Lagrangian on the vector field in the kinetic energy with respect to first order perturbation of the flow. This requires adjustment by the Lie bracket of vector field, given by operator
Defining the adjoint
meaning for all smooth
Computational anatomy is the study of the motions of submanifolds, points, curves, surfaces and volumes. Momentum associated to points, curves and surfaces are all singular, implying the momentum is concentrated on subsets of
Diffeomorphic shape momentum: a classical vector function
For the momentum density case
The Euler-Lagrange equation on diffeomorphisms, classically defined for momentum densities first appeared in for medical image analysis.
Riemannian exponential (geodesic positioning) and Riemannian logarithm (geodesic coordinates)
Global positioning systems based on systems of satellites provides a spatial navigation sytstem on the globe allowing electronic receivers to determine their location in the 3-dimensional coordinate system of longitude, latitude, and altitude to within meter scale.
In Medical imaging and Computational anatomy, positioning and coordinatizing shapes are fundamental operations; the system for positioning anatomical coordinates and shapes built on the metric and the Euler-Lagrange equation a geodesic positioning system as first explicated in Miller Trouve and Younes. Solving the geodesic from the initial condition
The Riemannian exponential satisfies
Computing the flow
Extended to the entire group they become
These are inverses of each other for unique solutions of Logarithm; the first is called geodesic positioning, the latter geodesic coordinates (see Exponential map, Riemannian geometry for the finite dimensional version).The geodesic metric is a local flattening of the Riemannian coordinate system (see figure).
Hamiltonian formulation of computational anatomy
In Computational anatomy the diffeomorphisms are used to push the coordinate systems, and the vector fields are used as the control within the anatomical orbit or morphological space. The model is that of a dynamical system, the flow of coordinates
This function is the extended Hamiltonian. The Pontryagin maximum principle gives the optimizing vector field which determines the geodesic flow satisfying
The Lagrange multiplier in its action as a linear form has its own inner product of the canonical momentum acting on the velocity of the flow which is dependent on the shape, e.g. for landmarks a sum, for surfaces a surface integral, and. for volumes it is a volume integral with respect to
with dynamics of canonical momentum reparameterizing the vector field along the geodesic
Stationarity of the Hamiltonian and kinetic energy along Euler–Lagrange
Whereas the vector fields are extended across the entire background space of
The geodesics connecting coordinate systems satisfying EL-General have stationarity of the Lagrangian. The Hamiltonian is given by the extremum along the path
The stationarity of the Hamiltonian demonstrates the interpretation of the Lagrange multiplier as momentum; integrated against velocity
The metric on geodesic flows of landmarks, surfaces, and volumes within the orbit
In Computational anatomy the submanifolds are pointsets, curves, surfaces and subvolumes which are the basic primitive forming the index sets or background space of medically imaged human anatomy. The geodesic flows of the submanifolds such as the landmarks, surface and subvolumes and the distance as measured by the geodesic flows of such coordinates, form the basic measuring and transporting tools of diffeomorphometry.
What is so important about the RKHS norm defining the kinetic energy in the action principle is that the vector fields of the geodesic motions of the submanifolds are superpositions of Green's Kernel's. For landmarks the superposition is a sum of weight kernels weighted by the canonical momentum which determines the inner product, for surfaces it is a surface integral, and for dense volumes it is a volume integral.
At
Landmark and surface submanifolds have Lagrange multiplier associated to a sum and surface integral, respectively; dense volumes an integral with respect to Lebesgue measure.
Landmark or pointset geodesics
For Landmarks, the Hamiltonian momentum is defined on the indices,
Surface geodesics
For surfaces, the Hamiltonian momentum is defined across the surface with the inner product
Volume geodesics
For volumes the Hamiltonian momentum is
Conservation laws on diffeomorphic shape momentum for computational anatomy
Given the least-action there is a natural definition of momentum associated to generalized coordinates; the quantity acting against velocity gives energy. The field has studied two forms, the momentum associated to the Eulerian vector field termed Eulerian diffeomorphic shape momentum, and the momentum associated to the initial coordinates or canonical coordinates termed canonical diffeomorphic shape momentum. Each has a conservation law.The conservation of momentum goes hand in hand with the EL-General. In Computational anatomy,
Conservation of Eulerian shape momentum was shown in and follows from EL-General; conservation of canonical momentum was shown in
Geodesic interpolation of information between coordinate systems via variational problems
Construction of diffeomorphic correspondences between shapes calculates the initial vector field coordinates
Matching based on minimizing kinetic energy action with endpoint condition
Conservation from EL-General extends the B.C. at
The running cost is reduced to the initial cost determined by
Matching based on geodesic shooting
The matching problem explicitly indexed to initial condition
Dense image matching in computational anatomy
Dense image matching has a long history now with the earliest efforts exploiting a small deformation framework. Large deformations began in the early 90's, with the first existence to solutions to the variational problem for flows of diffeomorphisms for dense image matching established in. Beg solved via one of the earliest LDDMM algorithms based on solving the variational matching with endpoint defined by the dense imagery with respect to the vector fields, taking variations with respect to the vector fields. Another solution for dense image matching reparameterizes the optimization problem in terms of the state
LDDMM dense image matching
For Beg's LDDMM, denote the Image
gives the variational problem
Beg's iterative LDDMM algorithm has fixed points which satisfy the necessary optimizer conditions. The iterative algorithm is given in Beg's LDDMM algorithm for dense image matching.
Hamiltonian LDDMM in the reduced advected state
Denote the Image
Viallard's iterative Hamiltonian LDDMM has fixed points which satisfy the necessary optimizer conditions.
Diffusion tensor image matching in computational anatomy
Dense LDDMM tensor matching takes the images as 3x1 vectors and 3x3 tensors solving the variational problem matching between coordinate system based on the principle eigenvectors of the diffusion tensor MRI image (DTI) denoted
Coordinate system transformation based on DTI imaging has exploited two actions one based on the principle eigen-vector or entire matrix.
LDDMM matching based on the principal eigenvector of the diffusion tensor matrix takes the image
LDDMM matching based on the entire tensor matrix has group action becomes
The variational problem matching onto the principal eigenvector or the matrix is described LDDMM Tensor Image Matching.
High Angular Resolution Diffusion Image (HARDI) matching in computational anatomy
High angular resolution diffusion imaging (HARDI) addresses the well-known limitation of DTI, that is, DTI can only reveal one dominant fiber orientation at each location. HARDI measures diffusion along
Dense LDDMM ODF matching takes the HARDI data as ODF at each voxel and solves the LDDMM variational problem in the space of ODF. In the field of information geometry, the space of ODF forms a Riemannian manifold with the Fisher-Rao metric. For the purpose of LDDMM ODF mapping, the square-root representation is chosen because it is one of the most efficient representations found to date as the various Riemannian operations, such as geodesics, exponential maps, and logarithm maps, are available in closed form. In the following, denote square-root ODF (
where
Based on this metric of ODF, we define a variational problem assuming that two ODF volumes can be generated from one to another via flows of diffeomorphisms
where
The LDDMM variational problem is defined as
This group action of diffeomorphisms on ODF reorients the ODF and reflects changes in both the magnitude of
This LDDMM-ODF mapping algorithm has been widely used to study brain white matter degeneration in aging, Alzheimer's disease, and vascular dementia. The brain white matter atlas generated based on ODF is constructed via Bayesian estimation. Regression analysis on ODF is developed in the ODF manifold space in.
Metamorphosis
The principle mode of variation represented by the orbit model is change of coordinates. For setting in which pairs of images are not related by diffeomorphisms but have photometric variation or image variation not represented by the template, active appearance modelling has been introduced, originally by Edwards-Cootes-Taylor and in 3D medical imaging in. In the context of Computational Anatomy in which metrics on the anatomical orbit has been studied, metamorphosis for modelling structures such as tumors and photometric changes which are not resident in the template was introduced in for Magnetic Resonance image models, with many subsequent developments extending the metamorphosis framework.
For image matching the image metamorphosis framework enlarges the action so that
Then the matching problem takes a form with equality boundary conditions:
Matching landmarks, curves, surfaces
Transforming coordinate systems based on Landmark point or fiducial marker features dates back to Bookstein's early work on small deformation spline methods for interpolating correspondences defined by fiducial points to the two-dimensional or three-dimensional background space in which the fiducials are defined. Large deformation landmark methods came on in the late 90's. The above Figure depicts a series of landmarks associated three brain structures, the amygdala, entorhinal cortex, and hippocampus.
Matching geometrical objects like unlabelled point distributions, curves or surfaces is another common problem in Computational Anatomy. Even in the discrete setting where these are commonly given as vertices with meshes, there are no predetermined correspondences between points as opposed to the situation of landmarks described above. From the theoretical point of view, while any submanifold
Landmark or point matching with correspondence
Denoted the landmarked shape
The geodesic Eulerian momentum is a generalized function
The iterative algorithm for large deformation diffeomorphic metric mapping for landmarks is given.
Measure matching: unregistered landmarks
Glaunes and co-workers first introduced diffeomorphic matching of pointsets in the general setting of matching distributions. As opposed to landmarks, this includes in particular the situation of weighted point clouds with no predefined correspondences and possibly different cardinalities. The template and target discrete point clouds are represented as two weighted sums of Diracs
The matching problem between a template and target point cloud may be then formulated using this kernel metric for the endpoint matching term:
where
Curve matching
In the one dimensional case, a curve in 3D can be represented by an embedding
In the situation of oriented curves, currents give an efficient setting to construct invariant matching terms. In such representation, curves are interpreted as elements of a functional space dual to the space vector fields, and compared through kernel norms on these spaces. Matching of two curves
with the endpoint term
the derivative
Varifold is an alternative to currents when orientation becomes an issue as for instance in situations involving multiple bundles of curves for which no "consistent" orientation may be defined. Varifolds directly extend 0-dimensional measures by adding an extra tangent space direction to the position of points, leading to represent curves as measures on the product of
where
Surface matching
Surface matching share many similarities with the case of curves. Surfaces in
Oriented surfaces can be represented as 2-currents which are dual to differential 2-forms. In
with the endpoint term
with
This surface mapping algorithm has been validated for brain cortical surfaces against CARET and FreeSurfer. LDDMM mapping for multiscale surfaces is discussed in.
For non-orientable or non-oriented surfaces, the varifold framework is often more adequate. Identifying the parametric surface
where
Growth and atrophy from longitudinal time-series
There are many settings in which there are a series of measurements, a time-series to which the underlying coordinate systems will be matched and flowed onto. This occurs for example in the dynamic growth and atrophy models and motion tracking such as have been explored in An observed time sequence is given and the goal is to infer the time flow of geometric change of coordinates carrying the exemplars or templars through the period of observations.
The generic time-series matching problem considers the series of times is
There have been at least three solutions offered thus far, piecewise geodesic, principal geodesic and splines.
The random orbit model of computational anatomy
The random orbit model of Computational Anatomy first appeared in modelling the change in coordinates associated to the randomness of the group acting on the templates, which induces the randomness on the source of images in the anatomical orbit of shapes and forms and resulting observations through the medical imaging devices. Such a random orbit model in which randomness on the group induces randomness on the images was examined for the Special Euclidean Group for object recognition in.
Depicted in the figure is a depiction of the random orbits around each exemplar,
The random orbit model induces the prior on shapes and images
Shown in the Figure on the right the cartoon orbit, are a random spray of the subcortical manifolds generated by randomizing the vector fields
The Bayesian model of computational anatomy
The central statistical model of Computational Anatomy in the context of medical imaging has been the source-channel model of Shannon theory; the source is the deformable template of images
See The Bayesian model of computational anatomy for discussions (i) MAP estimation with multiple atlases, (ii) MAP segmentation with multiple atlases, MAP estimation of templates from populations.
Statistical shape theory in computational anatomy
Shape in computational anatomy is a local theory, indexing shapes and structures to templates to which they are bijectively mapped. Statistical shape in Computational Anatomy is the empirical study of diffeomorphic correspondences between populations and common template coordinate systems. Interestingly, this is a strong departure from Procrustes Analyses and shape theories pioneered by David G. Kendall in that the central group of Kendall's theories are the finite-dimensional Lie groups, whereas the theories of shape in Computational Anatomy have focused on the diffeomorphism group, which to first order via the Jacobian can be thought of as a field–thus infinite dimensional–of low-dimensional Lie groups of scale and rotations.
The random orbit model provides the natural setting to understand empirical shape and shape statistics within Computational anatomy since the non-linearity of the induced probability law on anatomical shapes and forms
Performing empirical statistics on this tangent space at the identity is the natural way for inducing probability laws on the statistics of shape. Since both the vector fields and the Eulerian momentum
This is depicted in the accompanying figure where sub-cortical brain structures are depicted in a two-dimensional coordinate system based on inner products of their initial vector fields that generate them from the template is shown in a 2-dimensional span of the Hilbert space.
Template estimation from populations
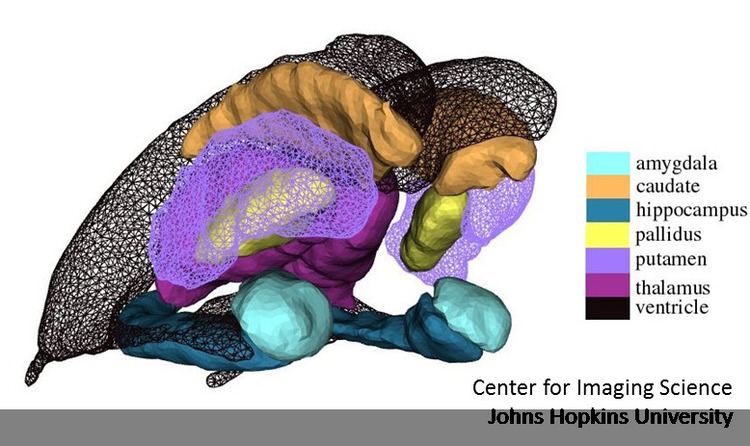
The study of shape and statistics in populations are local theories, indexing shapes and structures to templates to which they are bijectively mapped. Statistical shape is then the study of diffeomorphic correspondences relative to the template. A core operation is the generation of templates from populations, estimating a shape that is matched to the population. There are several important methods for generating templates including methods based on Frechet averaging, and statistical approaches based on the expectation-maximization algorithm and the Bayes Random orbit models of Computational anatomy. Shown in the accompanying figure is a subcortical template reconstruction from the population of MRI subjects.
Software for diffeomorphic mapping
Software suites containing a variety of diffeomorphic mapping algorithms include the following:
