Entrez 23597 | Ensembl ENSG00000123130 | |
 | ||
External IDs MGI: 1928940 HomoloGene: 8206 GeneCards: ACOT9 | ||
Acyl-CoA thioesterase 9 is a protein that is encoded by the human ACOT9 gene. It is a member of the acyl-CoA thioesterase superfamily, which is a group of enzymes that hydrolyze Coenzyme A esters. There is no known function, however it has been shown to act as a long-chain thioesterase at low concentrations, and a short-chain thioesterase at high concentrations.
Contents
Locus
The ACOT9 gene is located at p22.11 on chromosome X. Located on the minus strand of the chromosome, the start is at 23,721,777 bp and the end is at 23,761,407 bp, which is a span of 39,631 base pairs.
Aliases
ACOT9 gene is known primarily for encoding the Acyl-CoA thioesterase 9 protein. Other, less commonly used names for the gene are ACATE2, and MT-ACT48.
Function
The protein encoded by the ACOT9 gene is part of a family of Acyl-CoA thioesterases, which catalyze the hydrolysis of various Coenzyme A esters of various molecules to the free acid plus CoA. These enzymes have also been referred to in the literature as acyl-CoA hydrolases, acyl-CoA thioester hydrolases, and palmitoyl-CoA hydrolases. The reaction carried out by these enzymes is as follows:
CoA ester + H2O → free acid + coenzyme A
These enzymes use the same substrates as long-chain acyl-CoA synthetases, but have a unique purpose in that they generate the free acid and CoA, as opposed to long-chain acyl-CoA synthetases, which ligate fatty acids to CoA, to produce the CoA ester. The role of the ACOT- family of enzymes is not well understood; however, it has been suggested that they play a crucial role in regulating the intracellular levels of CoA esters, Coenzyme A, and free fatty acids. Recent studies have shown that Acyl-CoA esters have many more functions than simply an energy source. These functions include allosteric regulation of enzymes such as acetyl-CoA carboxylase, hexokinase IV, and the citrate condensing enzyme. Long-chain acyl-CoAs also regulate opening of ATP-sensitive potassium channels and activation of Calcium ATPases, thereby regulating insulin secretion. A number of other cellular events are also mediated via acyl-CoAs, for example signal transduction through protein kinase C, inhibition of retinoic acid-induced apoptosis, and involvement in budding and fusion of the endomembrane system. Acyl-CoAs also mediate protein targeting to various membranes and regulation of G Protein α subunits, because they are substrates for protein acylation. In the mitochondria, acyl-CoA esters are involved in the acylation of mitochondrial NAD+ dependent dehydrogenases; because these enzymes are responsible for amino acid catabolism, this acylation renders the whole process inactive. This mechanism may provide metabolic crosstalk and act to regulate the NADH/NAD+ ratio in order to maintain optimal mitochondrial beta oxidation of fatty acids. The role of CoA esters in lipid metabolism and numerous other intracellular processes are well defined, and thus it is hypothesized that ACOT- enzymes play a role in modulating the processes these metabolites are involved in.
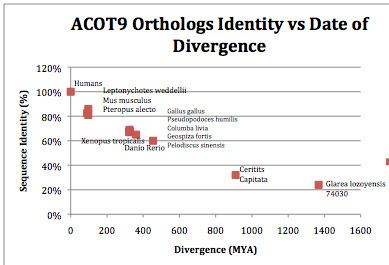
Orthologs
There are many orthologs of ACOT9, the house mouse (Mus musculus) being one of the most similar, where the ACOT9 gene is found at 72.38cM on chromosome X. The range of orthologs extends to mammals, birds, amphibians, anamorphic fungi, and others.
Paralogs
In mice, which is one of the closest orthologs, ACOT10 is a known paralog of the ACOT9 gene.
Expression
Expression of the ACOT9 is ubiquitous throughout the tissues in humans. Tissues with a value of over 500 in the large-scale analysis of the human transcriptome were the globus pallidus and colorectal adenocarcinoma. The expressed sequence tag (or EST) abundance profile also shows ubiquitous/near ubiquitous, expression throughout human tissues.
Transcription factors
There are numerous transcription factors throughout the ACOT9 promotor sequence. Some of the notable factors are heat shock factors and transcription factor II B (TFIIB) recognition elements.
Secondary structure
There are two regions in the ACOT9 gene sequence that are labeled as BFIT (Brown Fat Inducible Thioesterase) and BACH (Brain Acyl CoA Hydrolase) regions. These regions are part of a hotdog fold superfamily, which has been found to be used in a variety of cell roles. Predictions show there to be various alpha-helices throughout the structure, suggesting it is a transmembrane protein.
Interactions
A mitochondrial cleavage site can be found at amino acid 30 in the ACOT9 sequence, and the probability of export to the mitochondria is 0.9374. The Acyl-CoA thioesterase 9 protein is estimated to be 60.9% mitochondrial, 21.7% cytoplasmic, 8.7% nuclear, 4.3% in the plasma membrane, and 4.3% in the endoplasmic reticulum.
The ACOT9 protein has been found to interact with the following proteins either experimentally or through co-expression:
