Symbol U2 SO 0000392 | Rfam RF00004 Domain(s) Eukaryota | |
 | ||
GO 0000370 0045131 0005686 | ||
U2 spliceosomal RNA is a small nuclear RNA (snRNA) component of the spliceosome (involved in pre-mRNA splicing). Complementary binding between U2 snRNA (in an area lying towards the 5′ end but 3′ to hairpin I) and the branchpoint sequence (BPS) of the intron results in the bulging out of an unpaired adenosine, on the BPS, which initiates a nucleophilic attack at the intronic 5′ splice site, thus starting the first of two transesterification reactions that mediate splicing.
After recognition of the pre-mRNA 5′ splice site by U1 snRNA, U1 snRNA binds to this point. As soon as this bond forms, U2 snRNA binds to the branch point site, which forms a bulge at the branch point binding sequence. In addition, U6 snRNA associates with U2 snRNA and the 5′ splice site through base pairing interaction. The association of U2 snRNP with the pre-mRNA branch site plays a fundamental role during splicing assembly since it directly participates in chemical catalysis. Moreover, U2 snRNP undergoes several rearrangements along with the recognition of the branch point adenosine and formation of the base pair interaction between U2 snRNA and consensus sequence within the intron.
Contents
Structure and function
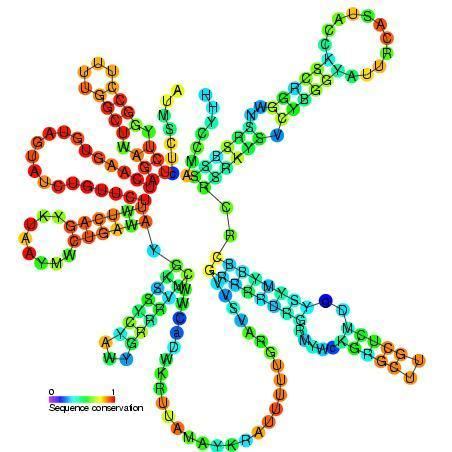
In humans, the U2 spliceosomal RNA is 187 base pairs long, forms five stem-loops, and possesses a 5′-trimethylguanosine five-prime cap. It is able to base-pair extensively with U6 spliceosomal RNA during the splicing process. Bases 33 to 38 are a conserved sequence that base-pairs with the adenosine branch site except for the adenosine itself, forcing the adenosine base to flip out and be exposed. Bases 99 to 105 form the Sm site, around which the heteroheptameric Sm ring is assembled, consisting of SmB/B′, SmD1/2/3, SmE, SmF, and SmG. U6-I and U6-II RNPs bind to stem-loop I. SF3a binds to stem loops IIa and IIb. U2 B″ binds to stem-loop IV, and U2 A′ interacts with U2 B″.
The 5′ end of U2 snRNA, which is highly conserved, contains several stem–loops, and single-stranded regions interact with U6 snRNA, the intron, and the snRNP proteins. Previous studies indicated that the stem I in 5′ terminus of the U2 snRNA has a function in early steps of spliceosome assembly. Structural studies of the U2-U6 complex show the presence of the U2 stem I as a stable structure in this complex.
NMR structures of U2 stem I from S. cerevisiae and the fully modified U2 stem I from humans reveal that there is a great similarity in structure; however, the stability of stem I in yeast and humans is extremely different.
Interactions
Interaction of the U2 snRNP to the branch site in humans occurs through the polypyrimidine tract in an ATP dependent manner. However, in yeast, introns do not contain a polypyrimidine tract at their 3′ splice site. In order for splicing to occur efficiently in yeast and mammals, it is necessary for U2 snRNA to base pair near the intron branch point.
U2 snRNA from yeast to mammals has been evolutionarily conserved. The spliceosome pathway in yeast extracts and mammalian extracts is different. A conserved intron branch sequence UACUAAC is required for U2 snRNP binding in yeast. However, the base paring interaction between U2 snRNA with the branch point of the intron and the pre-mRNA splicing mechanism is similar in both yeast and mammals.
