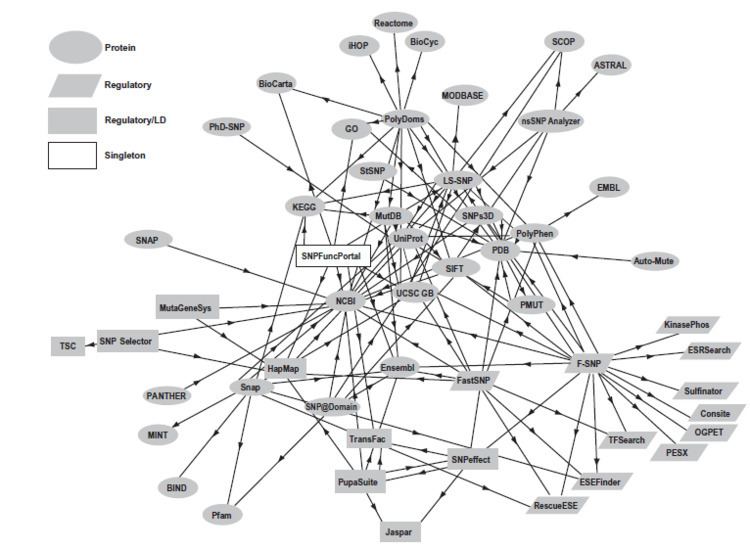
 | ||
Other subjects related | ||
Single nucleotide polymorphism (SNP) annotation is the process to predict the effect or function of an individual SNP using SNP annotational tools. In SNP annotation the biological information is extracted, collected and displayed in a clear form amenable to query. SNP functional annotation is done based on the available information on nucleic acid and protein sequence.
Contents
- Introduction
- SNP annotation evidence
- Gene based annotation
- Knowledge based annotation
- Functional annotation
- Transcriptional gene regulation
- Alternative splicing
- RNA processing and post transcriptional regulation
- Translation and post translational modifications
- Protein function
- Evolutionary conservation and nature selection
- List of available SNP annotation tools
- Algorithm used in annotation tools
- Comparison of variant annotation tools
- Conclusions
- References
Introduction
Single nucleotide polymorphism plays an important role in genome wide association studies because they act as primary biomarker. SNPs are currently the marker of choice due to their large numbers in virtually all populations of individuals. The location of these biomarkers can be tremendously important in terms of predicting functional significance, genetic mapping and population genetics. Each SNP represents a nucleotide change between two individuals at a defined location. SNPs are the most common genetic variant found in all individual with one SNP every 100–300 bp in some species. Since there is a massive number of SNPs on the genome, there is a clear need to prioritize SNPs according to their potential effect in order to expedite genotyping and analysis.
Annotating large numbers of SNPs is a difficult and complex process, which need computational methods to handle such a large dataset. Many tools available have been developed for SNP annotation in different organism, some of them are optimized for use with organisms densely sampled for SNPs (such as humans), but there are currently few tools available that are species non-specific or support non-model organism data. The majority of SNPs annotation tools provide computationally predicted putative deleterious effects of SNPs. These tools examine whether a SNP resides in functional genomic regions such as exons, splice sites, or transcription regulatory sites, and predict the potential corresponding functional effects that the SNP may have using a variety of machine-learning approaches. But the tools and systems that prioritize functionally significant SNPs, suffer from few limitations: First, they examine the putative deleterious effects of SNPs with respect to a single biological function that provide only partial information about the functional significance of SNPs. Second, current systems classify SNPs into deleterious or neutral group.
SNP annotation evidence
For SNP annotation many genetic and genomic information are used. Based on different feature used by the annotation tool, the SNP annotation can be classified into this category.
Gene based annotation
Genomic information from surrounding genomic elements is among the most useful information for interpreting the biological function of an observed variant. Information from a known gene is used as a reference to indicate whether the observed variant resides in or near a gene and if it has the potential to disrupt the protein sequence and its function. Gene based annotation is based on the fact that non-synonymous mutations can alter the protein sequence and that splice site mutation may disrupt the transcript splicing pattern.
Knowledge based annotation
Knowledge base annotation is done based on the information of gene attribute, protein function and its metabolism. In this type of annotation more emphasis is given to genetic variation that disrupts the protein function domain, protein-protein interaction and biological pathway. The non-coding region of genome contain many important regulatory elements including promoter, enhancer and insulator, any kind of change in this regulatory region can change the functionality of that protein. The mutation in DNA can change the RNA sequence and then influence the RNA secondary structure, RNA binding protein recognition and miRNA binding activity,.
Functional annotation
This method mainly identifies variant function based on the information whether the variant loci are in the known functional region that harbor genomic or epigenomic signals. The function of non-coding variants are extensive in terms of the affected genomic region and they involve in almost all processes of gene regulation from transcriptional to post translational level
Transcriptional gene regulation
Transcriptional gene regulation process depend on many spatial and temporal factor in the nucleus such as global or local chromatin states, nucleosome positioning, TF binding, enhancer/promoter activities. Variant that alter the function of any of these biological processes may alter the gene regulation and cause phenotypic abnormality. Genetic variants that located in distal regulatory region can affect the binding motif of TFs, chromatin regulators and other distal transcriptional factors, which disturb the interaction between enhancer/silencer and its target gene.
Alternative splicing
Alternative splicing is one of the most important components that show functional complexity of genome. Modified splicing has significant effect on the phenotype that is relevance to disease or drug metabolism. A change in splicing can be caused by modifying any of the components of the splicing machinery such as splice sites or splice enhancers or silencers. Modification in the alternative splicing site can lead to a different protein form which will show a different function. Humans use an estimated 100,000 different proteins or more, so some genes must be capable of coding for a lot more than just one protein. Alternative splicing occurs more frequently than was previously thought and can be hard to control; genes may produce tens of thousands of different transcripts, necessitating a new gene model for each alternative splice.
RNA processing and post transcriptional regulation
Mutations in the untranslated region (UTR) affect many post-transcriptional regulation. Distinctive structural features are required for many RNA molecules and cis-acting regulatory elements to execute effective functions during gene regulation. SNVs can alter the secondary structure of RNA molecules and then disrupt the proper folding of RNAs, such as tRNA/mRNA/lncRNA folding and miRNA binding recognition regions.
Translation and post translational modifications
Single nucleotide variant can also affect the cis-acting regulatory elements in mRNA’s to inhibit/promote the translation initiation. Change in the synonymous codons region due to mutation may affect the translation efficiency because of codon usage biases. The translation elongation can also be retarded by mutations along the ramp of ribosomal movement. In the post-translational level, genetic variants can contribute to proteostasis and amino acid modifications. However, mechanisms of variant effect in this field are complicated and there are only a few tools available to predict variant’s effect on translation related modifications.
Protein function
Non-synonymous is the variant in exons that change the amino acid sequence encoded by the gene, including single base changes and non frameshift indels. It has been extremely investigated the function of non-synonymous variants on protein and many algorithms have been developed to predict the deleteriousness and pathogenesis of single nucleotide variants (SNVs). Classical bioinformatics tools, such as SIFT, Polyphen and MutationTaster, successfully predict the functional consequence of non-synonymous substitution.
Evolutionary conservation and nature selection
Comparative genomics approaches were used to predict the function-relevant variants under the assumption that the functional genetic locus should be conserved across different species at an extensive phylogenetic distance. On the other hand, some adaptive traits and the population differences are driven by positive selections of advantageous variants, and these genetic mutations are functionally relevant to population specific phenotypes. Functional prediction of variants’ effect in different biological processes is pivotal to pinpoint the molecular mechanism of diseases/traits and direct the experimental validation.
List of available SNP annotation tools
To annotate large number of available NGS data, currently a large number of SNPs annotation tools is available. Some of them are specific to some specific SNPs annotation. Some of the available SNPs annotation tools are as follows SNPeff, VEP, ANNOVAR, FATHMM, PhD-SNP, PolyPhen-2, SuSPect, F-SNP, AnnTools, SeattleSeq, SNPit, SCAN, Snap, SNPs&GO, LS-SNP, Snat, TREAT, TRAMS, Maviant, MutationTaster, SNPdat, Snpranker, NGS – SNP, SVA, VARIANT, SIFT, PhD-SNP and FAST-SNP. Function and approach used in SNPs annotation tools are listed below
Algorithm used in annotation tools
Variant annotation tools use machine learning algorithms for prediction of variant. Different annotation tools use different algorithms. Common algorithms include:
Comparison of variant annotation tools
A large number of variant annotation tools are available for variant annotation but in some cases the prediction by the tools does not agree since the way the rules have been defined differ slightly between each application. It is frankly impossible to perform a perfect comparison of the tools. Not all the tools have same input and output and function. Here is a table of major annotation tools and it's functional area.
Source: S. Pabinger et al., 2012
Conclusions
The next generation of SNP annotation webservers can take advantage of the growing amount of data in core bioinformatics resources and use intelligent agents to fetch data from different sources as needed. From a user’s point of view, it is more efficient to submit a set of SNPs and receive results in a single step, which makes meta-servers the most attractive choice. However, if SNP annotation tools deliver heterogeneous data covering sequence, structure, regulation, pathways, etc., they must also provide frameworks for integrating data into a decision algorithm(s), and quantitative confidence measures so users can assess which data are relevant and which are not.
