Symbol SAND InterPro IPR000770 SUPERFAMILY 1h5p | Pfam PF01342 SCOP 1h5p Pfam structures | |
 | ||
In molecular biology, the protein domain SAND is named after a range of proteins in the protein family: Sp100, AIRE-1, NucP41/75, DEAF-1. It is localised in the cell nucleus and has an important function in chromatin-dependent transcriptional control. It is found solely in eukaryotes.
Contents
Function
The precise function of the protein domain SAND remains to be determined. Nevertheless, it is thought to be a DNA binding domain despite its beta structure. This function can be inferred by studying the DEAF-1 transcription factor. Here, the conserved positively charged residues in the SAND domains suggest the existence of negatively charged ligands. DNA is a negatively charged molecule due to the phosphate found in its backbone. Henceforth, this suggests that the SAND domain is the DNA-binding region of DEAF-1.
Structure
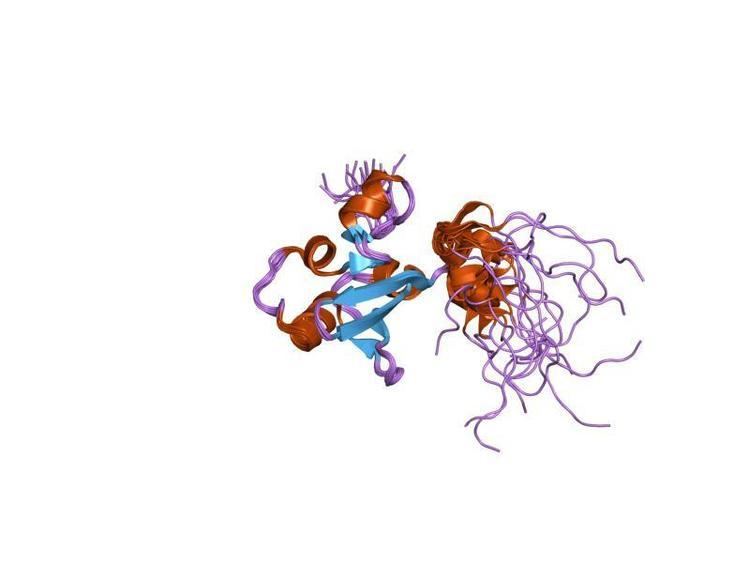
The structure of this protein domain contains a globular fold. It is thought to have an alpha/beta secondary structure that consists of five beta strands. This structure is made up of a five-stranded antiparallel beta-sheet with four alpha-helices. Further, the SAND domain is thought to have a modular structure; it can be associated with the bromodomain, the PHD finger and the MYND finger.
Conservation
This protein domain has a conserved region of around 80 residues. Mutations in this region lead to various human diseases, particularly in these proteins: Sp100 (Speckled protein 100 kDa), NUDR (Nuclear DEAF-1 related), GMEB (Glucocorticoid Modulatory Element Binding) proteins and AIRE-1 (Autoimmune regulator 1) proteins.
