 | ||
In molecular biology, a riboregulator is a ribonucleic acid (RNA) that responds to a signal nucleic acid molecule by Watson-Crick base pairing. A riboregulator may respond to a signal molecule in any number of manners including, translation (or repression of translation) of the RNA into a protein, activation of a ribozyme, release of silencing RNA (siRNA), conformational change, and/or binding other nucleic acids. Riboregulators contain two canonical domains, a sensor domain and an effector domain. These domains are also found on riboswitches, but unlike riboswitches, the sensor domain only binds complementary RNA or DNA strands as opposed to small molecules. Because binding is based on base-pairing, a riboregulator can be tailored to differentiate and respond to individual genetic sequences and combinations thereof.
Contents
Translational riboregulator
Translational riboregulators regulate the ability of a ribosome complex to scan, assemble, and/or translate an RNA molecule into a protein. In translational riboregulators, the RNA molecule is repressed or de-repressed depending on the secondary structure of the RNA molecule. Signal-responsive structures are usually introduced into the 5' untranslated region (5' UTR) of the RNA molecules using standard molecular biological techniques.
As discovered by Marilyn Kozak, the small (40S) ribosome complex scans an RNA molecule from 5' untranslated region to the start codon. When the complex encounters secondary structure, it must melt the structure to reach the start codon or it will fall off the molecule. The complex moves along through the untranslated region until it stalls just prior to reaching the start codon because it encounters a highly conserved sequence (a Kozak consensus sequence in eukaryotes, or Shine-Dalgarno sequence in prokaryotes). The stalled complex then combines with the large ribosome (60S) to begin translating the RNA into protein.
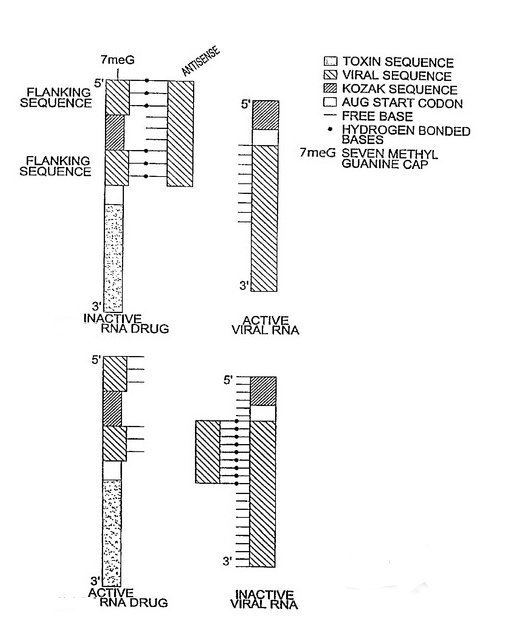
Lechner invented the first riboregulator in 1991. Lechner used a self pairing stem-loop which inhibited translation of a prokaryotic RNA, unless a complementary RNA sequence (anti-inhibitor) was present. In 1997, Black engineered the first eukaryotic riboregulator using antisense molecules to prevent translation (Nucline RNA). In the Nucline system, antisense molecules block translation unless removed via competitive hybridization and strand-displacement by specific signal RNA sequences such as HIV RNA and oncogene RNA. In 2003, Black demonstrated that Nucline RNA could perform Boolean and arithmetic operations (If-then-else, AND gate, OR gate and "Molar concentration of x > molar concentration of y" calculations ) in HIV infected monocytes and breast cancer cells.
In 2004, Isaacs et al. modified Lechner’s original system to demonstrate both trans and cis repression in a prokaryotic cells. Bayer & Smolke further developed a translational riboregulator in 2005 that could respond to small molecules creating a hybrid riboswitch/riboregulator molecule, termed an anti-switch. In an anti-switch, the presence of a small organic molecule binds an aptamer sequence in the RNA molecule which unmasks an otherwise sequestered antisense sequence, which can bind and block target RNA translation.
Ribozyme riboregulator
Ribozyme riboregulators regulate the ability of a catalytic RNA molecule to cleave a target nucleic acid sequence. In ribozyme riboregulators, a hammerhead ribozyme RNA molecule is activated or inactivated depending on the change of the secondary structure induced by hybridizing a signal molecule such as a cognate DNA or RNA sequence. In 2008, Win & Smolke designed a ribozyme regulator that could function in yeast cells that carried out Boolean operations similar to the earlier translational riboregulators, including AND, NAND, NOR, and OR gates.
RNAi-based riboregulator
RNAi riboregulators are small interfering RNAs which respond to a signal input such as complementary hybridization with a DNA or RNA molecule. The presence or absence of a target molecule determines whether the siRNA downregulates gene expression. In 2007, Rinaudo et al. demonstrated that RNAi based riboregulators can also perform Boolean operations in cells.
pH riboregulator
pH riboregulators regulate gene expression in response to pH changes. The only known pH riboregulator upregulates the alx gene in E. coli when in alkaline conditions, particularly above pH 8. The pH riboregulator was found, through experimentation, to reside in the 5' UTR of the alx gene. When in pH 7 or below, the pH riboregulator element (PRE) exists in an inactive 'N' state; in alkali conditions, RNA polymerase controls the folding of this ncRNA to its 'H' active form. The ribosome binding site of the alx gene is now exposed and the 30s subunit is allowed to bind.
Uses of riboregulators
Currently, riboregulators have found utility in the area of synthetic biology and personalized medicine. In synthetic biology, riboregulators can be used to regulate bacterial responses and probe gene regulatory networks. Undergraduate systems biology students use standardized riboregulators to compete in an annual bio-engineering competition (The international Genetically Engineered Machine competition, iGEM). In addition, because the various riboregulators can be tailored to respond to complex biological signals, riboregulators offer the promise of individually tailored RNA based drugs which respond to genes, small molecules and proteins in individuals cells.
