Entrez 5888 | Human Mouse Ensembl ENSG00000051180 | |
 | ||
Aliases RAD51, BRCC5, FANCR, HHsRad51, HsT16930, MRMV2, RAD51A, RECA, RAD51 recombinase External IDs OMIM: 179617 MGI: 97890 HomoloGene: 2155 GeneCards: RAD51 | ||
Doug bishop rad51 is a dmc1 accessory factor
RAD51 is a eukaryote gene. The protein encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA and yeast Rad51. The protein is highly conserved in most eukaryotes, from yeast to humans.
Contents
- Doug bishop rad51 is a dmc1 accessory factor
- Rad51 brca2 md binding
- Variants
- Family
- Function
- RAD51 expression in cancer
- RAD51 in double strand break repair
- MicroRNA control of RAD51 expression
- Pathology
- Interactions
- References
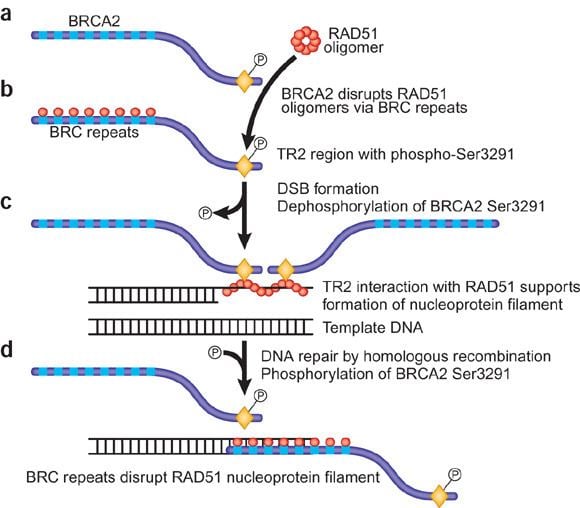
Rad51 brca2 md binding
Variants
Two alternatively spliced transcript variants of this gene, which encode distinct proteins, have been reported. Transcript variants utilizing alternative polyA signals exist.
Family
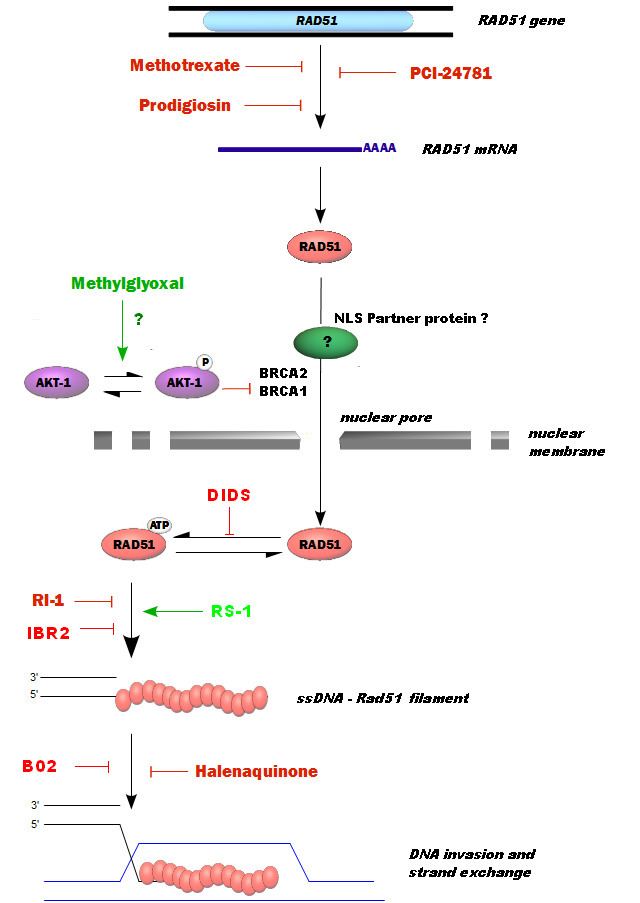
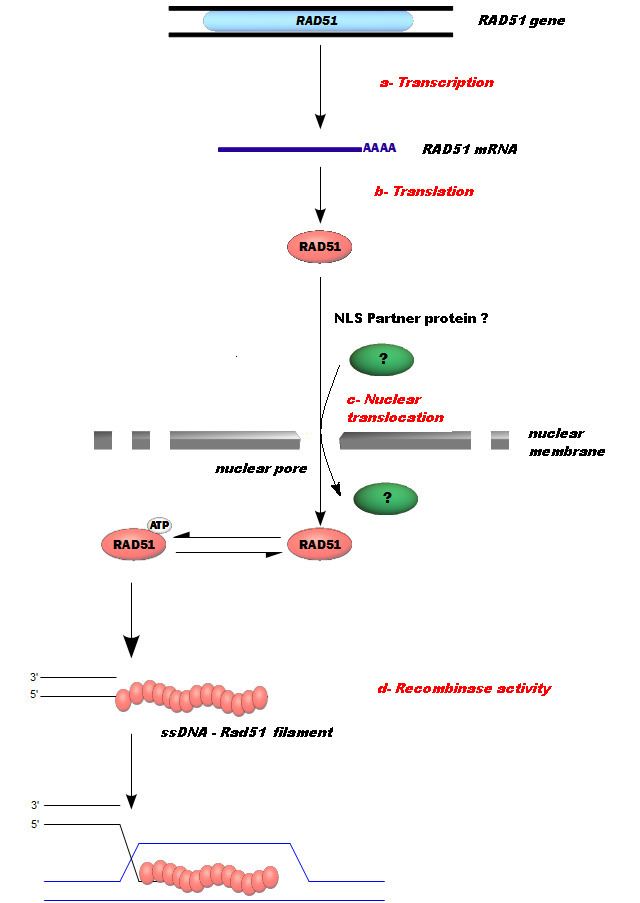
In mammals, seven recA-like genes have been identified: Rad51, Rad51L1/B, Rad51L2/C, Rad51L3/D, XRCC2, XRCC3, and DMC1/Lim15. All of these proteins, with the exception of meiosis-specific DMC1, are essential for development in mammals. Rad51 is a member of the RecA-like NTPases.
Function
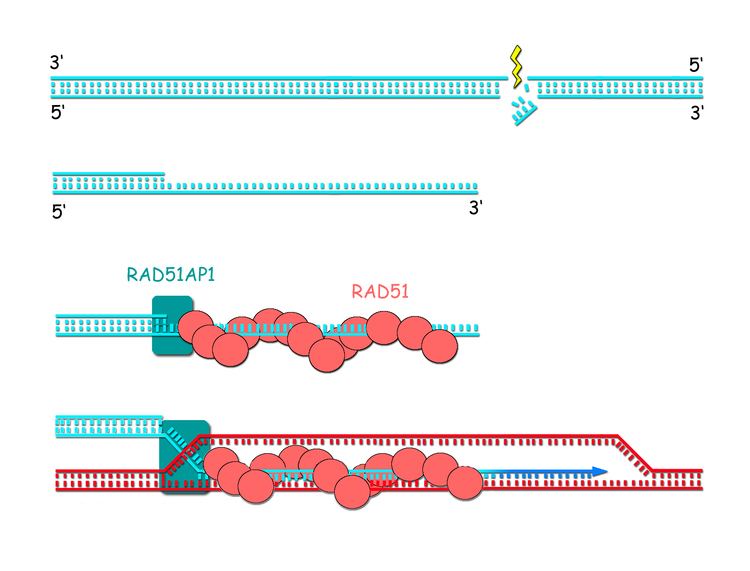
In humans, RAD51 is a 339-amino acid protein that plays a major role in homologous recombination of DNA during double strand break repair. In this process, an ATP dependent DNA strand exchange takes place in which a template strand invades base-paired strands of homologous DNA molecules. RAD51 is involved in the search for homology and strand pairing stages of the process.
Unlike other proteins involved in DNA metabolism, the RecA/Rad51 family forms a helical nucleoprotein filament on DNA.
This protein can interact with the ssDNA-binding protein RPA, BRCA2, PALB2 and RAD52.
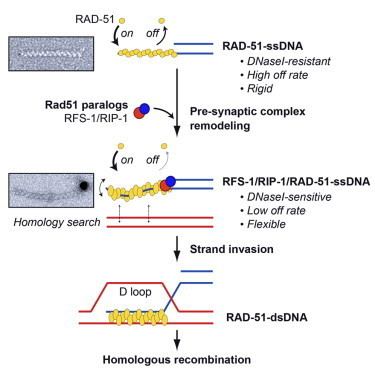
The structural basis for Rad51 filament formation and its functional mechanism still remain poorly understood. However, recent studies using fluorescent labeled Rad51 have indicated that Rad51 fragments elongate via multiple nucleation events followed by growth, with the total fragment terminating when it reaches about 2 μm in length. Disassociation of Rad51 from dsDNA, however, is slow and incomplete, suggesting that there is a separate mechanism that accomplishes this.
RAD51 expression in cancer
In eukaryotes, RAD51 protein has a central role in homologous recombinational repair. RAD51 catalyses strand transfer between a broken sequence and its undamaged homologue to allow re-synthesis of the damaged region (see homologous recombination models).
Numerous studies report that RAD51 is over-expressed in different cancers (see Table 1). In many of these studies, elevated expression of RAD51 is correlated with decreased patient survival. There are also some reports of under-expression of RAD51 in cancers (see Table 1).
Where RAD51 expression was measured in conjunction with BRCA1 expression, an inverse correlation was found. This was interpreted as selection for increased RAD51 expression and thus increased homologous recombinational repair (HRR) (by the HRR RAD52-RAD51 back-up pathway) to compensate for the added DNA damages remaining when BRCA1 was deficient.
Many cancers have epigenetic deficiencies in various DNA repair genes (see Frequencies of epimutations in DNA repair genes in cancers), likely causing increased unrepaired DNA damages. The over expression of RAD51 seen in many cancers may reflect compensatory RAD51 over expression (as in BRCA1 deficiency) and increased HRR to at least partially deal with such excess DNA damages.
Under-expression of RAD51 would itself lead to increased unrepaired DNA damages. Replication errors past these damages (see translesion synthesis), would lead to increased mutations and cancer.
RAD51 in double-strand break repair
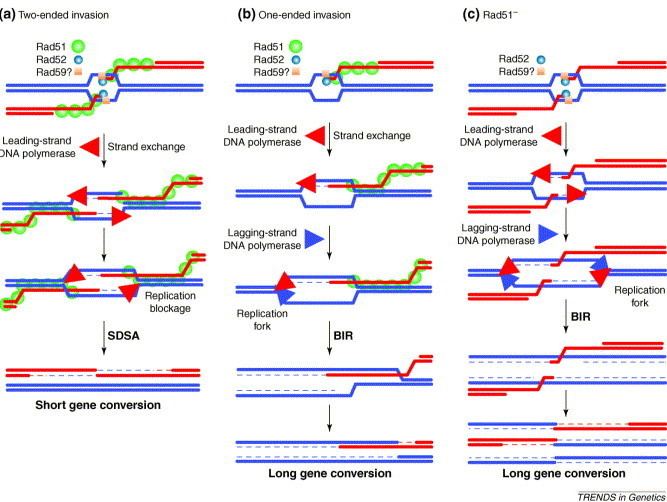
Double-strand break (DSB) repair by homologous recombination is initiated by 5' to 3' strand resection (DSB resection). In humans, the DNA2 nuclease cuts back the 5'-to-3' strand at the DSB to generate a 3' single-strand DNA overhang strand.
A complex of paralogs (see Figure) of RAD51 is essential for RAD51 protein recruitment or stabilization at damage sites in eukaryotes.
In eukaryotes, five paralogs of RAD51 are expressed in somatic cells, including RAD51B (RAD51L1), RAD51C (RAD51L2), RAD51D (RAD51L3), XRCC2 and XRCC3. They each share about 25% amino acid sequence identity with RAD51 and with each other.
The RAD51 paralogs are all required for efficient DNA double-strand break repair by homologous recombination and depletion of any paralog results in significant decreases in homologous recombination frequency.
The paralogs form two distinct complexes: BCDX2 (RAD51B-RAD51C-RAD51D-XRCC2) and CX3 (RAD51C-XRCC3). These two complexes act at two different stages of homologous recombinational DNA repair. The BCDX2 complex is responsible for RAD51 recruitment or stabilization at damage sites. The BCDX2 complex appears to act by facilitating the assembly or stability of the RAD51 nucleoprotein filament. The CX3 complex acts downstream of RAD51 recruitment to damage sites.
Another complex, the BRCA1-PALB2-BRCA2 complex, and the RAD51 paralogs cooperate to load RAD51 onto ssDNA coated with RPA to form the essential recombination intermediate, the RAD51-ssDNA filament.
In mice and humans, the BRCA2 complex primarily mediates orderly assembly of RAD51 on ssDNA, the form that is active for homologous pairing and strand invasion. BRCA2 also redirects RAD51 from dsDNA and prevents dissociation from ssDNA. However, in the presence of a BRCA2 mutation, human RAD52 can mediate RAD51 assembly on ssDNA and substitute for BRCA2 in homologous recombinational DNA repair, though with lower efficiency than BRCA2.
Further steps are detailed in the article Homologous recombination.
MicroRNA control of RAD51 expression
In mammals, microRNAs (miRNAs) regulate about 60% of the transcriptional activity of protein-encoding genes. Some miRNAs also undergo methylation-associated silencing in cancer cells. If a repressive miRNA is silenced by hypermethylation or deletion, then a gene it is targeting becomes over-expressed.
At least eight miRNAs have been identified that repress RAD51 expression, and five of them appear to be important in cancer. For instance, in triple negative breast cancers (TNBC), over-expression of miR-155 occurs together with repression of RAD51. Further tests directly showed that transfecting breast cancer cells with a vector over-expressing miR-155 represses RAD51, causing decreased homologous recombination and increased sensitivity to ionizing radiation.
Four further miRNAs that repress RAD51 (miR-148b* and miR-193b*, miR-506, and miR-34a) are under-expressed in cancers, presumably leading to induction of RAD51.
Under-expression of miR-148b* and miR-193b* cause an observed induction of RAD51 expression. Deletions of 148b* and miR-193b* in serous ovarian tumors correlate with increased incidences of (possibly carcinogenic) losses of heterozygosity (LOH). This excess LOH was thought to be due to excess recombination caused by induced expression of RAD51.
Under-expression of miR-506 is associated with early time to recurrence (and reduced survival) for epithelial ovarian cancer patients.
Methylation of the promoter of miR-34a, resulting in under-expression of miR-34a, is observed in 79% of prostate cancers and 63% of primary melanomas. Under-expressed levels of miR-34a are also seen in 63% of non-small cell lung cancers, and 36% of colon cancers. miR-34a is also generally under-expressed in primary neuroblastoma tumors.
Table 2 summarizes these five microRNAs, their over or under expression, and the cancers in which their altered expression was noted to occur.
The information summarized in Table 2 suggests that under-expression of microRNAs (causing induction of RAD51) occurs frequently in cancers. Over-expression of a microRNA that causes repression of RAD51 appears to be less frequent. The data in Table 1 (above) indicates that, in general, over-expression of RAD51 is more frequent in cancers than under-expression.
Three other microRNAs were identified, by various criteria, as likely to repress RAD51 (miR-96, miR-203, and miR-103/107). These microRNAs were then tested by over-expressing them in cells in vitro, and they were found to indeed repress RAD51. This repression was generally associated with decreased HR and increased sensitivity of the cells to DNA damaging agents.
Pathology
This protein is also found to interact with PALB2 and BRCA2, which may be important for the cellular response to DNA damage. BRCA2 is shown to regulate both the intracellular localization and DNA-binding ability of this protein. Loss of these controls following BRCA2 inactivation may be a key event leading to genomic instability and tumorigenesis.
Several alterations of the Rad51 gene have been associated with an increased risk of developing breast cancer. The breast cancer susceptibility protein BRCA2 and PALB2 controls the function of Rad51 in the pathway for DNA repair by homologous recombination. In addition to the data listed in Table 1, increased RAD51 expression levels have been identified in metastatic canine mammary carcinoma, indicating that genomic instability plays an important role in the carcinogenesis of this tumor type.
Interactions
RAD51 has been shown to interact with:
