RNA type Cis-regulatory element | Rfam RF01752 | |
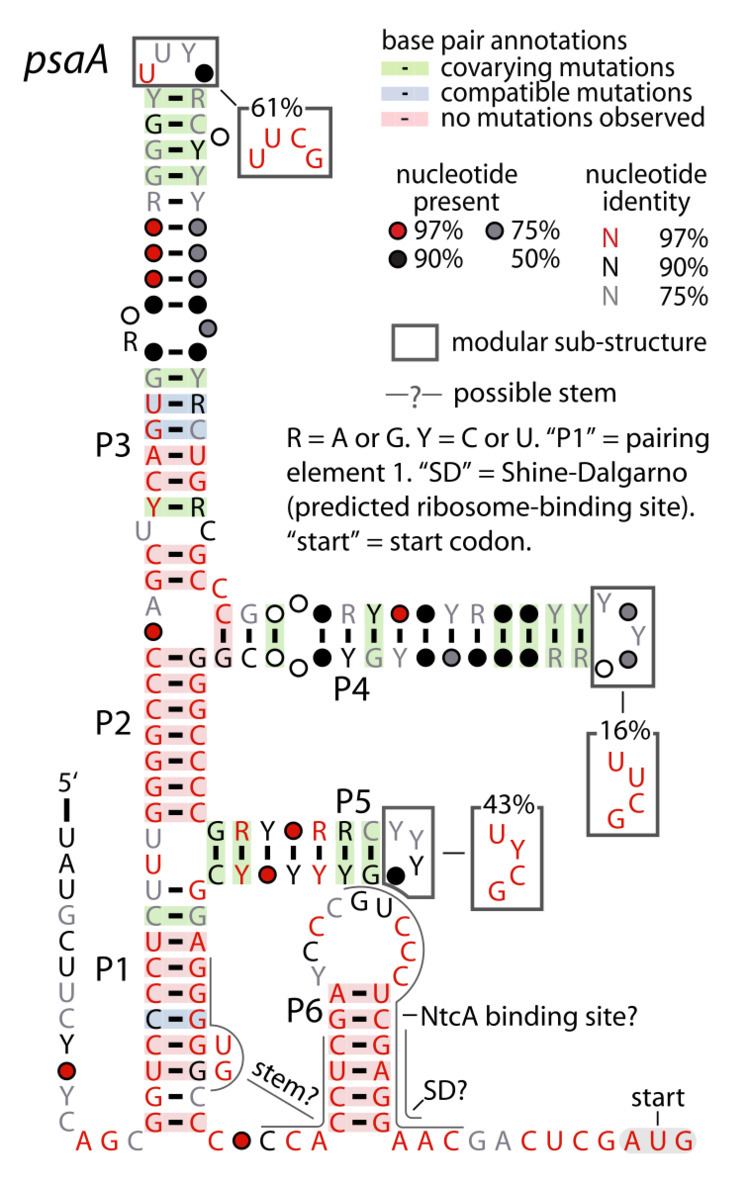 | ||
The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.
psaAB genes are known to be regulated in species of cyanobacteria that do not use psaA RNAs, and this system of regulation involves transcription factor proteins. In this system, the expression of psaAB genes is increased when cells are growing with limited amount of light, presumably to maximize their energy from the limited resource. On the other hand, the genes' expression is reduced when light levels are high, presumable to reduce damage that can be caused by free radicals that are a byproduct of photosynthesis. A region of the psaA RNA motif corresponds to the expected sequence of a binding site of the NtcA protein, which is involved in nitrogen regulation.
Structurally, the psaA RNA motif consists of a somewhat complex arrangement of stems resulting from base pairing, and many stem-loops are terminated by the stable UNCG tetraloop. Most of the conserved nucleotides in the motif participate in standard Watson-Crick base pairs. However, since the precise function of psaA RNAs is unknown, it is also unknown what biochemical role these features play.
