EC number 5.2.1.8 ExPASy NiceZyme view Gene ontology 0003755 | CAS number 95076-93-0 | |
 | ||
Medical vocabulary what does peptidylprolyl isomerase mean
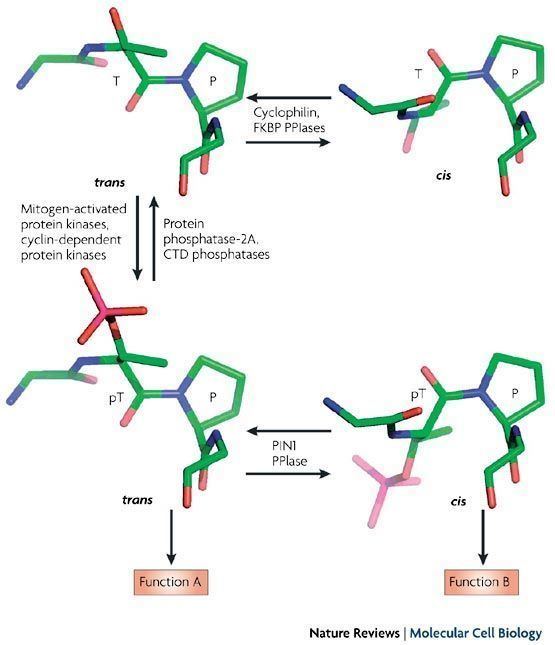
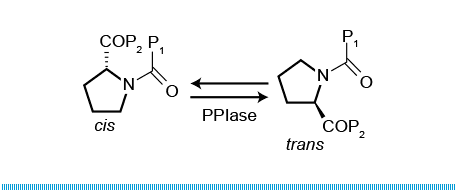
Prolyl isomerase (also known as peptidylprolyl isomerase or PPIase) is an enzyme (EC 5.2.1.8) found in both prokaryotes and eukaryotes that interconverts the cis and trans isomers of peptide bonds with the amino acid proline. Proline has an unusually conformationally restrained peptide bond due to its cyclic structure with its side chain bonded to its secondary amine nitrogen. Most amino acids have a strong energetic preference for the trans peptide bond conformation due to steric hindrance, but proline's unusual structure stabilizes the cis form so that both isomers are populated under biologically relevant conditions. Proteins with prolyl isomerase activity include cyclophilin, FKBPs, and parvulin, although larger proteins can also contain prolyl isomerase domains.
Contents
- Medical vocabulary what does peptidylprolyl isomerase mean
- Protein folding
- Evidence for proline isomerization
- Assays for prolyl isomerase activity
- References
Protein folding
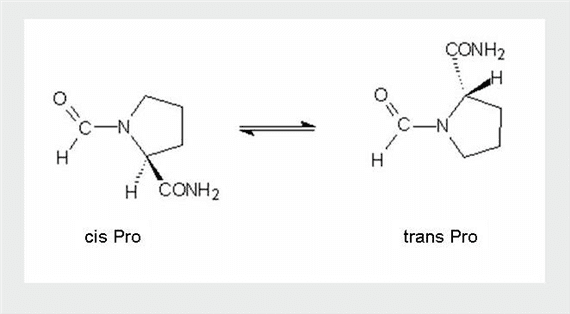
Proline is unique among the natural amino acids in having a relatively small difference in free energy between the cis configuration of its peptide bond and the more common trans form. The activation energy required to catalyse the isomerisation between cis and trans is relatively high: ~20kcal/mol (c.f. ~0kcal/mol for regular peptide bonds). Unlike regular peptide bonds, the X-prolyl peptide bond will not adopt the intended conformation spontaneously, thus, the process of cis-trans isomerization can be the rate-limiting step in the process of protein folding. Prolyl isomerases therefore function as protein folding chaperones. Cis peptide bonds N-terminal to proline residues are often located at the first residue of certain types of tight turns in the protein backbone. Proteins that contain structural cis-prolines in the native state include ribonuclease A, ribonuclease T1, beta lactamase, cyclophilin, and some interleukins.
Prolyl isomerase folding can be autocatalytic and therefore the speed of folding depends on reactant concentration. Parvulin and human cytosolic FKBP are thought to catalyze their own folding processes.
Evidence for proline isomerization
Methods for identifying the presence of a rate-limiting proline isomerization process in a protein folding event include:

- Activation energies consistent with proline isomerization, which typically has an activation of about 20 kcal/mol.
- Two-state folding kinetics indicative of both fast-folding and slow-folding populations in the unfolded or denatured state.
- "Double-jump" assays in which proline-containing proteins are unfolded and refolded, and the population of non-native proline conformations are studied as a function of the extent of folding.
- Acceleration of the in vitro folding rate by the addition of a prolyl isomerase.
- Acceleration of the in vitro folding rate in mutant protein variants with one or more proline residues replaced by another amino acid.
It is important to note that not every proline peptide bond is critical to the structure or function of a protein, and not every such bond has a significant influence on folding kinetics, especially trans bonds. Furthermore, some prolyl isomerases have a degree of sequence specificity and therefore may not catalyze the isomerization of prolines in certain sequence contexts.
Assays for prolyl isomerase activity
Prolyl isomerase activity was first discovered using a chymotrypsin-based assay. The proteolytic enzyme chymotrypsin has a very high substrate specificity for the four-residue peptide Ala-Ala-Pro-Phe only when the proline peptide bond is in the trans state. Adding chymotrypsin to a solution containing a reporter peptide with this sequence results in the rapid cleavage of about 90% of the peptides, while those peptides with cis proline bonds - about 10% in aqueous solution - are cleaved at a rate limited by uncatalyzed proline isomerization. The addition of a potential prolyl isomerase will accelerate this latter reaction phase if it has true prolyl isomerase activity.