Other names Peptide chip | ||
 | ||
Uses To study binding properties and functionality and kinetics of protein-protein interactions | ||
Peptide microarray service
A peptide microarray (also commonly known as peptide chip or peptide epitope microarray) is a collection of peptides displayed on a solid surface, usually a glass or plastic chip. Peptide chips are used by scientists in biology, medicine and pharmacology to study binding properties and functionality and kinetics of protein-protein interactions in general. In basic research, peptide microarrays are often used to profile an enzyme (like kinase, phosphatase, protease, acetyltransferase, histone deacetylase etc.), to map an antibody epitope or to find key residues for protein binding. Practical applications are seromarker discovery, profiling of changing humoral immune responses of individual patients during disease progression, monitoring of therapeutic interventions, patient stratification and development of diagnostic tools and vaccines.
Contents
- Peptide microarray service
- Peptide microarray applications
- Principle
- Production of a peptide microarray
- Applications of peptide microarrays
- Immunology
- Enzyme profiling
- Analysis and evaluation of results
- References
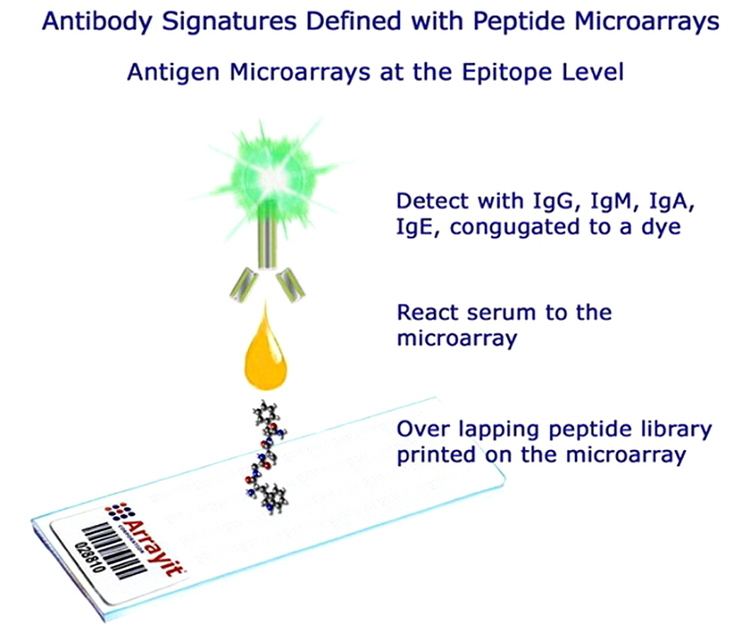
Peptide microarray applications
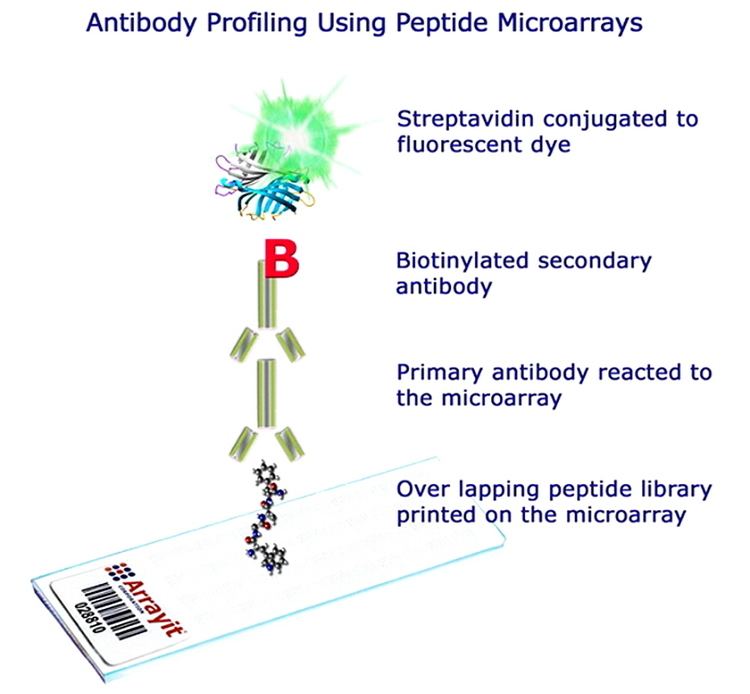
Principle
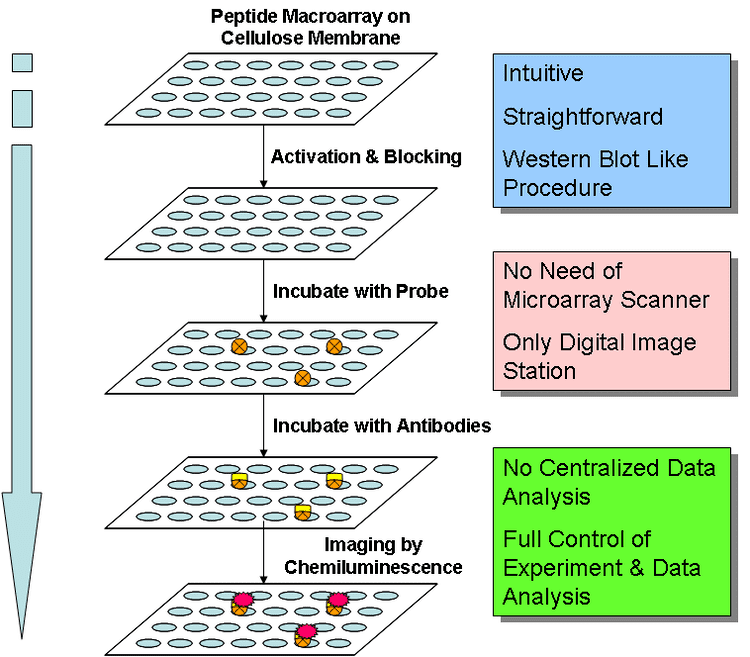
The assay principle of peptide microarrays is similar to an ELISA protocol. The peptides (up to tens of thousands in several copies) are linked to the surface of a glass chip typically the size and shape of a microscope slide. This peptide chip can directly be incubated with a variety of different biological samples like purified enzymes or antibodies, patient or animal sera, cell lysates etc. After several washing steps a secondary antibody with the needed specificity (e.g. anti IgG human/mouse or anti phosphotyrosine or anti myc) is applied. Usually, the secondary antibody is tagged by a fluorescence label that can be detected by a fluorescence scanner. Other detection methods are chemiluminescence, colorimetric or autoradiography.
Peptide microarrays show several advantages over protein microarrays:
Production of a peptide microarray
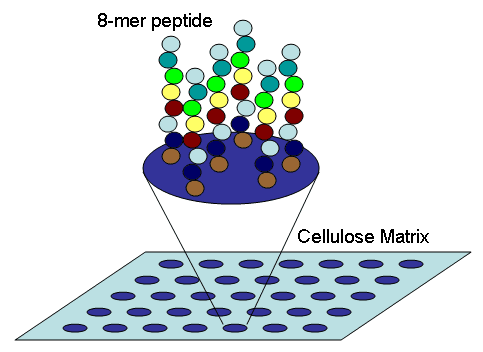
A peptide microarray is a planar slide with peptides spotted onto it or assembled directly on the surface by in-situ synthesis. Whereas peptides spotted can undergo quality controls that include mass spectrometer analysis and concentration normalization before spotting and result from a single synthetic batch, peptides synthesized directly on the surface may suffer from batch-to-batch variation and limited quality control options. However, peptide synthesis on chip allows the parallel synthesis of tenths of thousands of peptides providing larger peptide libraries paired with lower synthesis costs. Peptides are ideally covalently linked through an chemoselective bond leading to peptides with the same orientation for interaction profiling. Alternative procedures describe unspecific covalent binding and adhesive immobilization.
Applications of peptide microarrays

Peptide microarrays can be used to study different kinds of protein-protein interactions, specially those involving modular protein substructures called peptide recognition modules or, most communly, protein interaction domains. The reason for this is that such protein substructures recognize short linear motifs often exposed in natively unstructured regions of the binding partner, such that the interaction can be modelled in vitro by peptides as probes and the peptide recognition module as analyte. Most publications can be found in the context of immune monitoring and enzyme profiling.
Immunology
Enzyme profiling
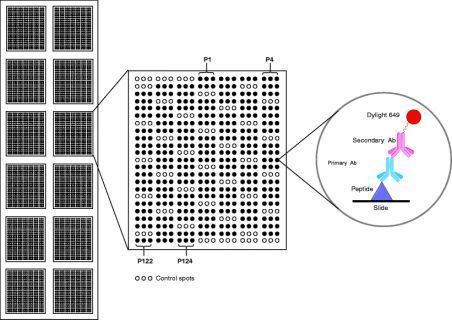
Analysis and evaluation of results
Data analysis and evaluation of results is the most important part of every microarray experiment. After scanning the microarray slides, the scanner records a 20-bit, 16-bit or 8-bit numeric image in tagged image file format (*.tif). The .tif-image enables interpretation and quantification of each fluorescent spot on the scanned microarray slide. This quantitative data is the basis for performing statistical analysis on measured binding events or peptide modifications on the microarray slide. For evaluation and interpretation of detected signals an allocation of the peptide spot (visible in the image) and the corresponding peptide sequence has to be performed. The data for allocation is usually saved in the GenePix Array List (.gal) file and supplied together with the peptide microarray. The .gal-file (a tab-separated text file) can be opened using microarray quantification software-modules or processed with a text editor (e.g. notepad) or Microsoft Excel. This "gal" file is most often provided by the microarray manufacturer and is generated by input txt files and tracking software built into the robots that do the microarray manufacturing.
