 | ||
In molecular biology a pan-genome (or supra-genome) describes the full complement of genes in a clade (typically applied to species in bacteria and archaea), which can have large variation in gene content among closely related strains. It is the union of the gene sets of all the strains of a clade (e.g. species). The significance of the pan-genome arises in an evolutionary context, especially with relevance to metagenomics, but is also used in a broader genomics context.
Contents
The pan-genome includes the "core genome" containing genes present in all strains, a "dispensable genome" containing genes present in two or more strains, and finally "unique genes" specific to single strains. However, these distinctions are not completely objective, since they depend on which genomes are included in the analysis.
Examples
The original pan-genome concept was developed by Tettelin et al. when they sequenced six strains of Streptococcus agalactiae which could be described as a core genome shared by all isolates, accounting for approximately 80% of any single genome, plus a dispensable genome consisting of partially shared and strain-specific genes. Extrapolation suggested that the gene reservoir in the S. agalactiae pan-genome is vast and that new unique genes will continue to be identified even after sequencing hundreds of genomes.
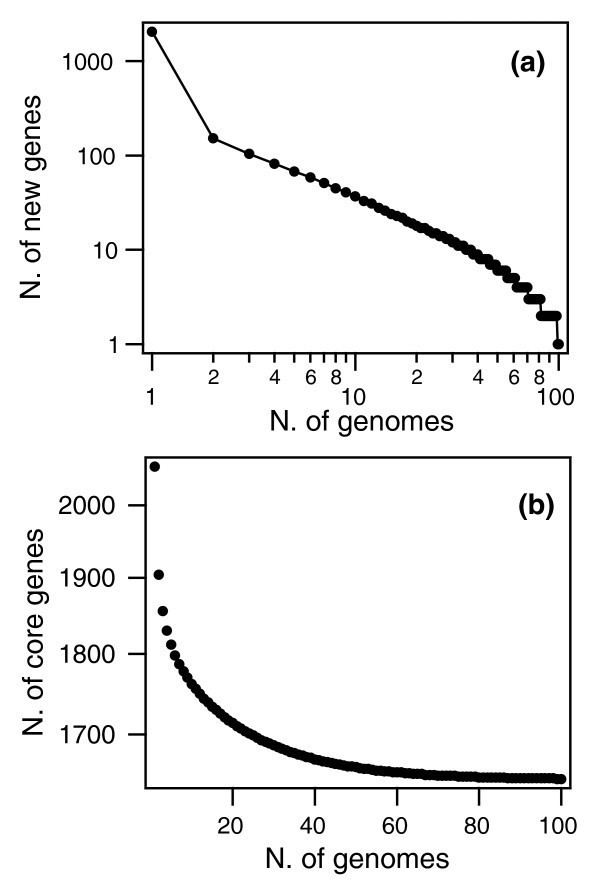
A similar pattern was found in Streptococcus pneumoniae when 44 strains were sequenced (see figure). With each new genome sequenced fewer new genes were discovered. In fact, the predicted number of new genes dropped to zero when the number of genomes exceeds 50 (note, however, that this is not a pattern found in all species). The main source of new genes in S. pneumoniae was Streptococcus mitis from which genes were transferred horizontally. The pan-genome size of S. pneumoniae increased logarithmically with the number of strains and linearly with the number of polymorphic sites of the sampled genomes, suggesting that acquired genes accumulate proportionately to the age of clones.
Another example for the latter can be seen in a comparison of the sizes of the core and the pan-genome of Prochlorococcus. The core genome set is logically much smaller than the pan-genome, which is used by different ecotypes of Prochlorococcus. A 2015 study on Prevotella bacteria isolated from humans, compared the gene repertoires of its species derived from different body sites of human. It also reported an open pan- genome showing vast diversity of gene pool.
The same group has developed a BPGA - A Pan-Genome Analysis Pipeline for prokaryotic genomes in 2016, which provides faster alternative over other software.
Software Tools
As interest in pangenomes increased, there have been a number of software tools developed to help analyze this kind of data. In 2015, a group reviewed the different kinds of analyses and tools a researcher may want to use. There are seven kinds of analyses software has been developed for to analyze pangenomes: cluster homologous genes; identify SNPs; plot pangenomic profiles; build phylogenetic relationships of orthologous genes/families of strains/isolates; function-based searching; annotation and/or curation; and visualizations.
The two most cited software tools at the end of 2014 were Panseq and the pan-genomes analysis pipeline (PGAP).
