Symbol N/A InterPro IPR009003 SUPERFAMILY 50494 | Pfam CL0124 SCOP 50494 Pfam structures | |
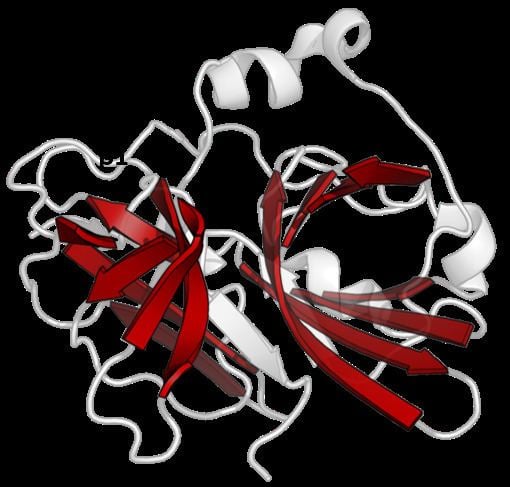 | ||
The PA clan (Proteases of mixed nucleophile, superfamily A) is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but sequence identity of <10%. The clan contains both cysteine and serine proteases (different nucleophiles). PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.
Contents
The PA clan represents an example of convergent evolution to the use of a catalytic triad for hydrolysis. It also is an example of extreme divergent evolution of active sites in enzymes.
History
In the 1960s, the sequence similarity of several proteases indicated that they were evolutionarily related. These were grouped into the chymotrypsin-like serine proteases (now called the S1 family). As the structures of these, and other proteases were solved by X-ray crystallography in the 1970s and 80s, it was noticed that several viral proteases such as Tobacco Etch Virus protease showed structural homology despite no discernible sequence similarity and even a different nucleophile. Based on structural homology, a superfamily was defined and later named the PA clan (by the MEROPS classification system). As more structures are solved, more protease families have been added to the PA clan superfamily.
Etymology
The P refers to Proteases of mixed nucleophile. The A indicates that it was the first such clan to be identified (there also exist the PB, PC, PD and PE clans).
Structure
Despite retaining as little as 10% sequence identity, PA clan members isolated from viruses, prokaryotes and eukaryotes show structural homology and can be aligned by structural similarity (e.g. with DALI).
Double β-barrel
PA clan proteases all share a core motif of two β-barrels with covalent catalysis performed by an acid-histidine-nucleophile catalytic triad motif. The barrels are arranged perpendicularly beside each other with hydrophobic residues holding them together as the core scaffold for the enzyme. The triad residues are split between the two barrels so that catalysis takes place at their interface.
Viral protease loop
In addition to the double β-barrel core, some viral proteases (such as TEV protease) have a long, flexible C-terminal loop that forms a lid to which completely covers the substrate and create a binding tunnel. This tunnel contains a set of tight binding pockets such that each side chain of the substrate peptide (P6 to P1’) is bound in a complementary site (S6 to S1’) and specificity is endowed by the large contact area between enzyme and substrate. Conversely, cellular proteases that lack this loop, such as trypsin have broader specificity.
Evolution and function
The structural homology described above indicates that the PA clan members are descended from a common ancestor of the same fold. Although PA clan proteases share a common ancestor some families use serine as the nucleophile in their catalytic triad whilst others use cysteine. The catalytic triad is a set of three residues that together perform nucleophilic catalysis and are therefore a vital component of the enzyme active site. This is therefore an extreme example of divergent enzyme evolution since during evolutionary history, the core catalytic residue of the enzyme has switched. In addition to their structural similarity, directed evolution has been shown to be able to convert a cysteine protease into an active serine protease. All cellular PA clan proteases are serine proteases, however there are both serine and cysteine protease families of viral proteases.
In addition to divergence in their core catalytic machinery, the PA clan proteases also show wide divergent evolution in function. Members of the PA clan can be found in eukaryotes, prokaryotes and viruses and encompass a wide range of functions. In mammals, some are involved in blood clotting (e.g. thrombin) and so have high substrate specificity as well as digestion (e.g. trypsin) with broad substrate specificity. Several snake venoms are also PA clan proteases, such as pit viper haemotoxin and interfere with the victim's blood clotting cascade. Additionally, bacteria such as Staphylococcus aureus secrete exfoliative toxin which digest and damage the host's tissues. Finally, many viruses express their genome as a single, massive polyprotein and use a protease to cleave this into functional units (e.g. polio, norovirus, and TEV proteases).
Families
Within the PA clan (P=proteases of mixed nucleophiles), families are designated by their catalytic nucleophile (C=cysteine proteases, S=serine proteases). Despite the lack of sequence homology for the PA clan as a whole, individual families within it can be identified by sequence similarity.
