 | ||
N6-Methyladenosine (m6A) is an abundant modification in mRNA and is found within some viruses, and most eukaryotes including mammals, insects, plants and yeast. It is also found in tRNA, rRNA, and small nuclear RNA (snRNA) as well as several long non-coding RNA, such as Xist.
Contents

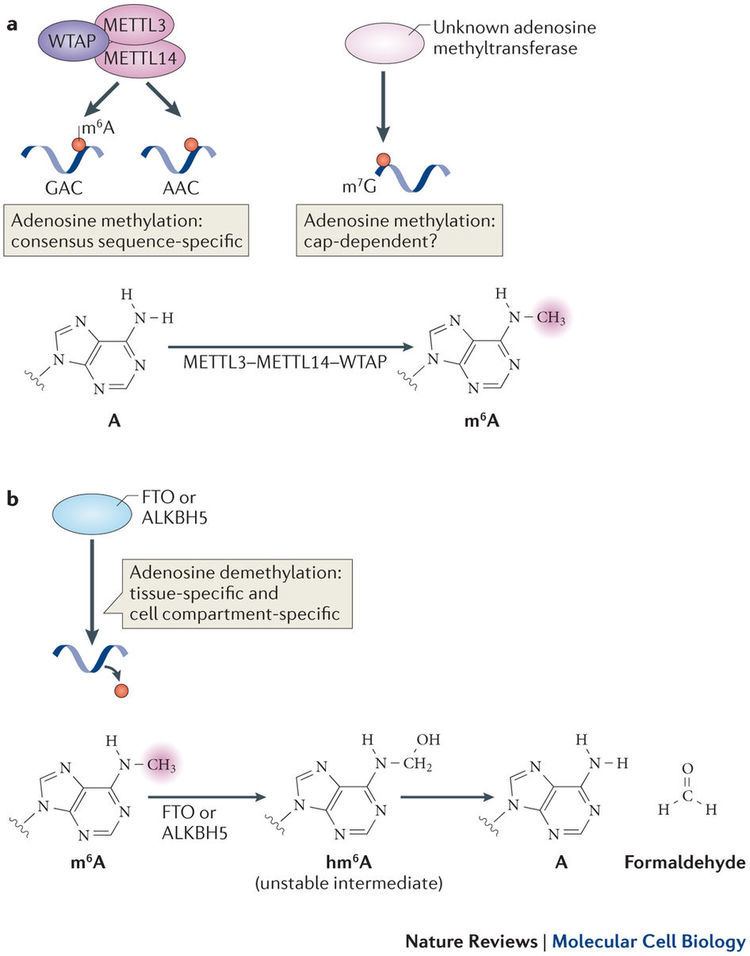
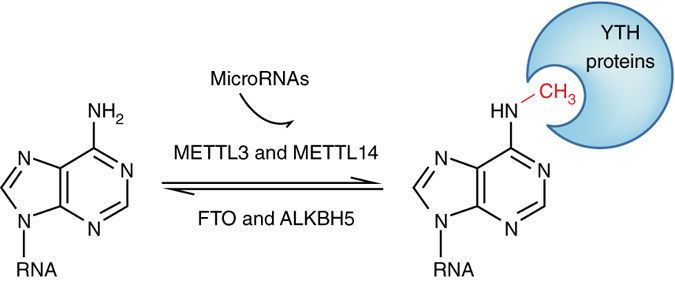
Adenosine methylation is directed by a large m6A methyltransferase complex containing METTL3 as the SAM-binding sub-unit. In vitro, this methyltransferase complex preferentially methylates RNA oligonucleotides containing GGACU and a similar preference was identified in vivo in mapped m6A sites in Rous sarcoma virus genomic RNA and in bovine prolactin mRNA. More recent studies have characterized other key components of the m6A methyltransferase complex in mammals, including METTL14, Wilms tumor 1 associated protein (WTAP) and KIAA1429. Following a 2010 speculation of m6A in mRNA being dynamic and reversible, the discovery of the first m6A demethylase, fat mass and obesity-associated protein (FTO) in 2011 confirmed this hypothesis and revitalized the interests in the study of m6A. A second m6A demethylase alkB homolog 5 (ALKBH5) was later discovered as well.
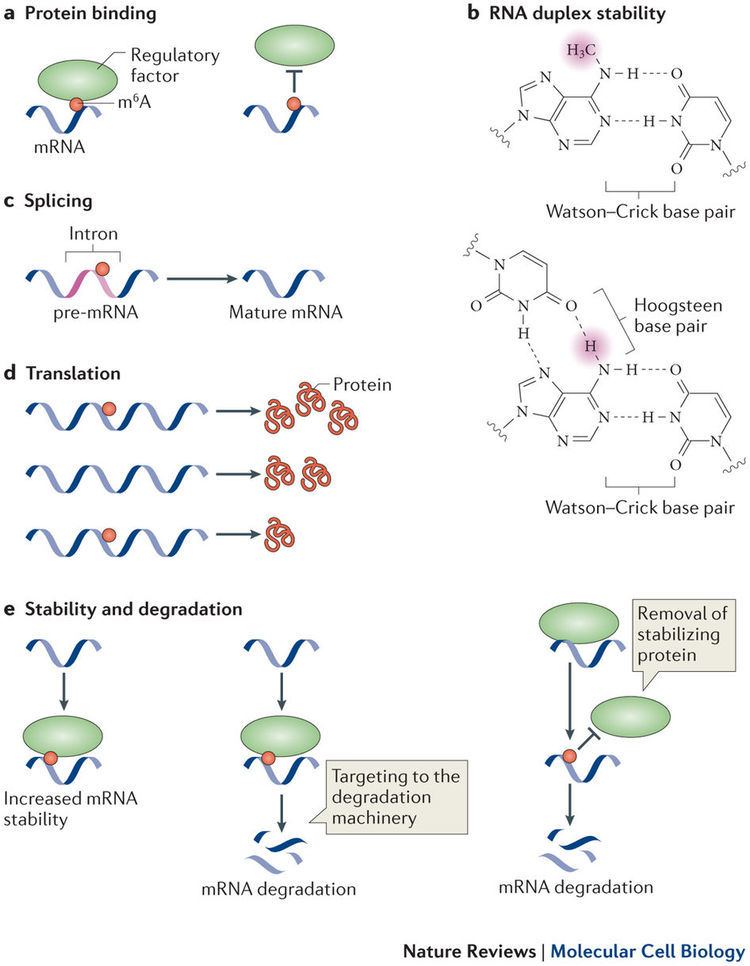
The biological functions of m6A are mediated through a group of RNA binding proteins that specifically recognize the methylated adenosine on RNA. These binding proteins are named m6A readers. The YT521-B homology (YTH) domain family of proteins (YTHDF1, YTHDF2, YTHDF3 and YTHDC1) have been characterized as direct m6A readers and have a conserved m6A-binding pocket. These m6A readers, together with m6A methyltransferases (writers) and demethylases (erasers), establish a complex mechanism of m6A regulation in which writers and erasers determine the distributions of m6A on RNA, whereas readers mediate m6A-dependent functions. m6A has also been shown to mediate a structural switch termed m6A switch.
Yeast
In budding yeast (Sacharomyces cerevisiae), the homologue of METTL3, IME4 is induced in diploid cells in response to nitrogen and fermentable carbon source starvation and is required for mRNA methylation and the initiation of correct meiosis and sporulation. mRNAs of IME1 and IME2, key early regulators of meiosis, are known to be targets for methylation, as are transcripts of IME4 itself.
Plants
In plants, the majority of the m6A is found within 150 nucleotides before the start of the poly(A) tail.
Mutations of MTA, the Arabidopsis thaliana homologue of METTL3, results in embryo arrest at the globular stage. A >90% reduction of m6A levels in mature plants leads to dramatically altered growth patterns and floral homeotic abnormalities.
Mammals
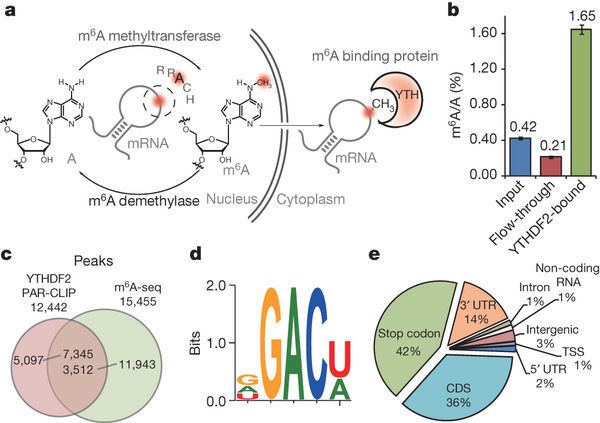
Mapping of m6A in human and mouse RNA has identified over 18,000 m6A sites in the transcripts of more than 7,000 human genes with a consensus sequence of [G/A/U][G>A]m6AC[U>A/C] consistent with the previously identified motif. The localization of individual m6A sites in many mRNAs is highly similar between human and mouse, and transcriptome-wide analysis reveals that m6A is found in regions of high evolutionary conservation. m6A is found within long internal exons and is preferentially enriched within 3’ UTRs and around stop codons. m6A within 3’ UTRs is also associated with the presence of microRNA binding sites; roughly 2/3 of the mRNAs which contain an m6A site within their 3’ UTR also have at least one microRNA binding site. By integrating all m6A sequencing data, a novel database called RMBase has identified and provided ~200,000 N6-Methyladenosines (m6A) sites in the human and mouse genomes.
Precise m6A mapping by m6A-CLIP/IP (briefly m6A-CLIP, in multiple tissues/cultured cells of mouse and human) revealed that a majority of m6A locates in the last exon of mRNAs, and the m6A enrichment around stop codons is a coincidence that many stop codons locate round the start of last exons where m6A is truly enriched. The major presence of m6A in last exon (>=70%) allows the potential for 3'UTR regulation, including alternative polyadenylation.
m6A is susceptible to dynamic regulation both throughout development and in response to cellular stimuli. Analysis of m6A in mouse brain RNA reveals that m6A levels are low during embryonic development and increase dramatically by adulthood. Additionally, silencing the m6A methyltransferase significantly affects gene expression and alternative RNA splicing patterns, resulting in modulation of the p53 (also known as TP53) signalling pathway and apoptosis.
The importance of m6A methylation for physiological processes was recently demonstrated. Inhibition of m6A methylation via pharmacological inhibition of cellular methylations or more specifically by siRNA-mediated silencing of the m6A methylase Mettl3 led to the elongation of the circadian period. In contrast, overexpression of Mettl3 led to a shorter period. The mammalian circadian clock, composed of a transcription feedback loop tightly regulated to oscillate with a period of about 24 hours, is therefore extremely sensitive to perturbations in m6A-dependent RNA processing, likely due to the presence of m6A sites within clock gene transcripts.
Clinical significance
Considering the versatile functions of m6A in various physiological processes, it is thus not surprising to find links between m6A and numerous human diseases; many originated from mutations or single nucleotide polymorphisms (SNPs) of cognate factors of m6A. The linkages between m6A and numerous cancer types have been indicated in reports that include stomach cancer, prostate cancer, breast cancer, pancreatic cancer, kidney cancer, mesothelioma, sarcoma, and leukaemia. The impacts of m6A on cancer cell proliferation might be much more profound with more data emerging. The depletion of METTL3 is known to cause apoptosis of cancer cells and reduce invasiveness of cancer cells, while the activation of ALKBH5 by hypoxia was shown to cause cancer stem cell enrichment. m6A has also been indicated in the regulation of energy homeostasis and obesity, as FTO is a key regulatory gene for energy metabolism and obesity. SNPs of FTO have been shown to associate with body mass index in human populations and occurrence of obesity and diabetes. The influence of FTO on pre-adipocyte differentiation has been suggested. The connection between m6A and neuronal disorders has also been studied. For instance, neurodegenerative diseases may be affected by m6A as the cognate dopamine signalling was shown to be dependent on FTO and correct m6A methylation on key signalling transcripts. The mutations in HNRNPA2B1, a potential reader of m6A, have been known to cause neurodegeneration.
Additionally, m6A has been reported to impact viral infections. Many RNA viruses including SV40, adenovirus, herpes virus, Rous sarcoma virus, and influenza virus have been known to contain the internal m6A methylation on virus genomic RNA. Several more recent studies have revealed that m6A regulators govern the efficiency of infection and replication of RNA viruses such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), and Zika virus (ZIKV). These results suggest m6A and its cognate factors play crucial roles in regulating virus life cycle and host-viral interactions.
