EC number 4.1.3.30 ExPASy NiceZyme view | CAS number 57827-77-7 | |
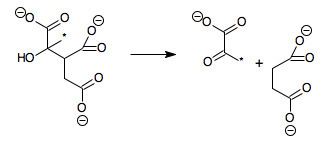 | ||
In enzymology, a methylisocitrate lyase (EC 4.1.3.30) is an enzyme that catalyzes the chemical reaction
Contents
(2S,3R)-3-hydroxybutane-1,2,3-tricarboxylateHence, this enzyme has one substrate, (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate (also known as 2-methylisocitrate), and two products, pyruvate and succinate.
The reaction is similar to that of isocitrate lyase, except that an additional methyl group (marked with an asterisk in the above scheme) is present, meaning that citrate is replaced by methylcitrate and glyoxylate by pyruvate.
This enzyme belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase (succinate-forming). Other names in common use include 2-methylisocitrate lyase, MICL, and (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase. This enzyme participates in propanoate metabolism.
Methylisocitrate lyase was discovered in 1976.
Structural studies
As of late 2007, 6 structures have been solved for this class of enzymes, with PDB accession codes 1MUM, 1O5Q, 1OQF, 1UJQ, 1XG3, and 1XG4. The structure is very similar to that of phosphoenolpyruvate mutase. A homotetrameric biological unit is composed of beta barrels with the active site at one end. A magnesium ion is present in the active site, and an active-site "gating loop" moves inward toward it when substrate binds and away with no substrate bound, thus shielding the reaction from solvent. Helices are present all around the beta barrels; in particular, a C-terminal helical domain splits off from the barrel to interact with the barrel of a neighboring subunit, in a "helix swapping" motif (see phosphoenolpyruvate mutase).
The following still shot from a ribbon kinemage shows one subunit from the crystal structure 1MUM, which includes a magnesium ion (gray) but no substrate; helices are red while loops are white and beta strands are green.
Biological function
Methylisocitrate lyase is used in the methylcitrate cycle, a modified version of the Krebs cycle that metabolizes propionyl coenzyme A instead of acetyl coenzyme A. The enzyme 2-methylcitrate synthase adds propionyl coenzyme A to oxaloacetate, yielding methylcitrate instead of citrate. But isomerizing methylcitrate to methylisocitrate and then subjecting it to MICL regenerates succinate, which proceeds as in the Krebs cycle, and pyruvate, which is easily metabolized by other pathways (e.g. decarboxylated to form acetyl coenzyme A and oxidized in the Krebs cycle). This allows catabolism of propionic acid—and, using beta oxidation, other fatty acids with odd numbers of carbons—without relying on coenzyme B12, a complex cofactor often used to metabolize propionate. The methylcitrate cycle is found in many microorganisms.
Methylisocitrate lyase plays a regulatory function in this cycle; it is activated by NAD but inhibited noncompetitively by NADH and NADPH.
