Ancestor L3 | ||
 | ||
Possible time of origin 60,000 years before present Descendants M1, M2, M3, M4'45, M5, M6, M7, M8, M9, M10'42, M12'G, M13, M14, M15, M21, M27, M28, M29'Q, M31'32, M33, M34, M35, M36, M39, M40, M41, M44, M46, M47'50, M48, M49, M51, D Defining mutations 263, 489, 10400, 14783, 15043 | ||
In human mitochondrial genetics, Haplogroup M is a human mitochondrial DNA (mtDNA) haplogroup. An enormous haplogroup spanning all the continents, the macro-haplogroup M, like its sibling N, is a descendant of haplogroup L3.
Contents
- Origins
- Haplogroup M1
- Haplogroup M23
- Asian origin hypothesis
- African origin hypothesis
- Dispersal
- Distribution
- Subgroups distribution
- Tree
- References
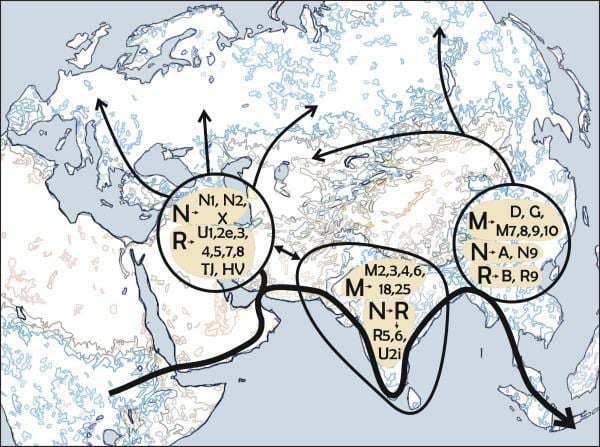
All mtDNA haplogroups considered native outside of Africa are descendants of either haplogroup M or its sibling haplogroup N. The geographical distributions of M and N are associated with discussions concerning out of Africa migrations and the subsequent colonization of the rest of the world. In particular, it is often taken to indicate that it is very likely that there was one particularly major prehistoric migration of humans out of Africa, and that both M and N were part of this colonization process.
Origins
There is a debate concerning geographical origins of Haplogroup M and its sibling haplogroup N. Both lineages are thought to have been the main surviving lineages involved in the out of Africa migration (or migrations) because all indigenous lineages found outside Africa belong to haplogroup M or haplogroup N. Yet to be conclusively determined is whether the mutations that define haplogroups M and N occurred in Africa before the exit from Africa or in Asia after the exit from Africa. Determining the origins of haplogroup M is further complicated by the fact that it is found in Africa and outside of Africa.
It is generally accepted that haplogroup M evolved shortly after the emergence of its parent clade haplogroup L3. Apart from haplogroup M and its sibling haplogroup N, the numerous other subclades of L3 are largely restricted to Africa, which suggests that L3 arose in Africa.
Haplogroup M1
Much of discussion concerning the origins of haplogroup M has been related to its subclade haplogroup M1, which is the only variant of macrohaplogroup M found in Africa. Two possibilities were being considered as potential explanations for the presence of M1 in Africa:
- M was present in the ancient population which later gave rise to both M1 in Africa, and M more generally found in Eurasia.
- The presence of M1 in Africa is the result of a back-migration from Asia which occurred sometime after the Out of Africa migration 40,000 years ago.
Haplogroup M23
In 2009, two independent publications reported a rare, deep-rooted subclade of haplogroup M, referred to as M23, that is present in Madagascar. The contemporary populations of Madagascar were formed in the last 2,000 years by the admixture of African and Southeast Asian (Austronesian) populations. M23 seems to be restricted to Madagascar as it was not detected anywhere else. M23 could have been brought to Madagascar from Asia where most deep rooted subclades of Haplogroup M are found.
Asian origin hypothesis
According to this theory, anatomically modern humans carrying ancestral haplogroup L3 lineages were involved in the Out of Africa migration from East Africa into Asia. Somewhere in Asia, the ancestral L3 lineages gave rise to haplogroups M and N. The ancestral L3 lineages were then lost by genetic drift as they are infrequent outside Africa. The hypothesis of Asia as the place of origin of macrohaplogroup M is supported by the following:
- The highest frequencies worldwide of macrohaplogroup M are observed in Asia, specifically in Bangladesh, China, India, Japan, Nepal, and Tibet, where frequencies range from 60%-80%. The total frequency of M subclades is even higher in some populations of Siberia or the Americas, but these small populations tend to exhibit strong genetic drift effects, and often their geographical neighbors exhibit very different frequencies.
- Deep time depth >50,000 years of western, central, southern and eastern Indian haplogroups M2, M38, M54, M58, M33, M6, M61, M62 and the distribution of macrohaplogroup M, do not rule out the possibility of macrohaplogroup M arising in Indian population.
- With the exception of the African specific M1, India has several M lineages that emerged directly from the root of haplogroup M.
- Only two subclades of haplogroup M, M1 and M23, are found in Africa, whereas numerous subclades are found outside Africa (with some discussion possible only about sub-clade M1, concerning which see below).
- Specifically concerning M1
African origin hypothesis
According to this theory, haplogroups M and N arose from L3 in an East African population that had been isolated from other African populations. Members of this population were involved in the out Africa migration and only carried M and N lineages. With the possible exception of haplogroup M1, all other M and N clades in Africa were lost by genetic drift.
The African origin of Haplogroup M is supported by the following arguments and evidence.
- L3, the parent clade of haplogroup M, is found throughout Africa, but is rare outside Africa. According to Toomas Kivisild (2003), "the lack of L3 lineages other than M and N in India and among non-African mitochondria in general suggests that the earliest migration(s) of modern humans already carried these two mtDNA ancestors, via a departure route over the Horn of Africa."
- Ancestral L3 lineages that gave rise to M and N have not been discovered outside Africa.
- Specifically concerning at least M1:
Dispersal
A number of studies have proposed that the ancestors of modern haplogroup M dispersed from Africa through the southern route across the Horn of Africa along the coastal regions of Asia onwards to New Guinea and Australia. These studies suggested that the migrations of haplogroups M and N occurred separately with haplogroup N heading northwards from East Africa to the Levant. However, the results of numerous recent studies indicate that there was only one migration out of Africa and that haplogroups M and N were part of the same migration. This is based on the analysis of a number of relict populations along the proposed beachcombing route from Africa to Australia, all of which possessed both haplogroups N and M.
A 2008 study by Abu-Amero et al., suggests that the Arabian Peninsula may have been the main route out of Africa. However, as the region lacks of autochthonous clades of haplogroups M and N the authors suggest that the area has been a more recent receptor of human migrations than an ancient demographic expansion center along the southern coastal route as proposed under the single migration Out-of-Africa scenario of the African origin hypothesis.
Distribution
M is the single most common mtDNA haplogroup in Asia, and peaks in Japan and Tibet, where it represents on average about 70% of the maternal lineages (160/216 = 74% Tibet, 205/282 = 73% Tōkai, 231/326 = 71% Okinawa, 148/211 = 70% Japanese, 50/72 = 69% Tibet, 150/217 = 69% Hokkaidō, 24/35 = 69% Zhongdian Tibetan, 175/256 = 68% northern Kyūshū, 38/56 = 68% Qinghai Tibetan, 16/24 = 67% Diqing Tibetan, 66/100 = 66% Miyazaki, 33/51 = 65% Ainu, 214/336 = 64% Tōhoku, 75/118 = 64% Tokyo (JPT)) and is ubiquitous in India and South Korea, where it has approximately 60% frequency. Among Chinese people both inside and outside of China, haplogroup M accounts for approximately 50% of all mtDNA on average, but the frequency varies from approximately 40% in Hans from Hunan and Fujian in southern China to approximately 60% in Shenyang, Liaoning in northeastern China.
Haplogroup M accounts for approximately 42% of all mtDNA in Filipinos, among whom it is represented mainly by M7c3c and E. In Vietnam, haplogroup M has been found in 37% (52/139) to 48% (20/42) of samples of Vietnamese and in 32% (54/168) of a sample of Chams from Bình Thuận Province. Haplogroup M accounts for 43% (92/214) of all mtDNA in a sample of Laotians, with its subclade M7 (M7b, M7c, and M7e) alone accounting for a full third of all haplogroup M, or 14.5% (31/214) of the total sample.
In Oceania, Haplogroup M has been found in 35% (17/48) of a sample of Papua New Guinea highlanders from the Bundi area and in 28% (9/32) of a sample of Aboriginal Australians from Kalumburu in northwestern Australia. A study published in 2008 found Haplogroup M in 42% (60/144) of a pool of samples from nine language groups in the Admiralty Islands of Papua New Guinea, of which 50 belonged to typically Near Oceanian subclades (Q1, Q2) and 10 belonged to typically East or Southeast Asian subclades (M7b, M7c1c, E1b). In a study published in 2015, Haplogroup M was found in 21% (18/86) of a sample of Fijians and in 0% (0/21) of a sample of Rotumans.
Haplogroup M is also relatively common in Northeast Africa, occurring especially among Somalis, Libyans and Oromos at frequencies over 20%. Toward the northwest, the lineage is found at comparable frequencies among the Tuareg in Mali and Burkina Faso, particularly the M1a2 subclade (18.42%).
Due to its great age, haplogroup M is an mtDNA lineage which does not correspond well to present-day ethnic groups. It is found among Siberian, Native American, East Asian, Southeast Asian, Central Asian, South Asian, Melanesian, European, Northeast African, and various Middle Eastern populations at varying frequencies.
Among the descendant lineages of haplogroup M are C, D, E, G, Q, and Z. Z and G are found in North Eurasian populations, C and D exists among North Eurasian and Native American populations, E is observed in Southeast Asian populations, and Q is common among Melanesian populations. The lineages M2, M3, M4, M5, M6, M18 and M25 are exclusive to South Asia, with M2 reported to be the oldest lineage on the Indian sub-continent.
In 2013, four ancient specimens dated to around 2,500 BC-500 AD, which were excavated from the Tell Ashara (Terqa) and Tell Masaikh (Kar-Assurnasirpal) archaeological sites in the Euphrates valley, were found to belong to mtDNA haplotypes associated with the M4b1, M49 and/or M61 haplogroups. Since these clades are not found among the current inhabitants of the area, they are believed to have been brought at a more remote period from east of Mesopotamia; possibly by either merchants or the founders of the ancient Terqa population.
In 2016, three Late Pleistocene European hunter-gatherers were also found to carry M lineages. Two of the specimens were from the Goyet archaeological site in Belgium and were dated to 34,000 and 35,000 years ago, respectively. The other ancient individual hailed from the La Rochette site in France and was dated to 28,000 years ago.
Subgroups distribution
Tree
This phylogenetic tree of haplogroup M subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation and subsequent published research.
