Ancestor C M130 | Defining mutations M217, P44, PK2 | |
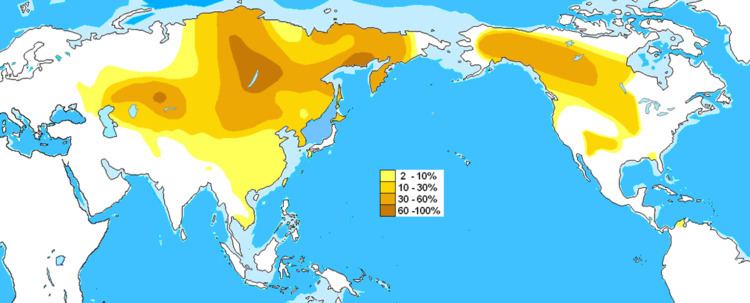 | ||
Possible time of origin 11,900 ± 4,800 years before present14,920 ± 3,830 years (evolutionary mutation rate) or 4,120 ± 1,060 years (genealogical mutation rate)34,200 [95% CI 31,800 <-> 36,600] ybp Descendants C-M93 (C2a); C-CTS117 (C2b); C-P53.1 (C2c); C-P62 (C2d); C-F2613/Z1338 (C2e) Highest frequencies Oroqen 61%-91%, Evens 5%-74%, Evenks 44%-71%, Buryats 7%-84%, Mongolians 51%-54%, Kazakhs 40%-60.7%, Tanana 42%, Hazaras 35% - 40%, Nivkhs 38%, Koryaks 33%, Daur 31%, Yukaghir 31%, Sibe 27%, Manchu 26%-27%, Altai 22%-24%, Hezhe 22%, Kyrgyz 20%, Uzbeks 20%, Tujia 18%, Hani 18%, Cheyenne 16%, Apache 15%, Tuvans 11% - 15%, Ainu 12.5%-25%, Koreans 6%-26%, Hui 11%, Sioux 11%, Nogais 14%, Crimean Tatars 9%, Han Chinese 0%-23.5%, Vietnamese 4.3%-12.5% | ||
Haplogroup C-M217, also known as C2 (and previously as C3), is a Y-chromosome DNA haplogroup. It is the most frequently occurring branch of the wider Haplogroup C (M130).
Contents
M217 is found at high frequencies among Central Asian peoples, indigenous Siberians, and some Native peoples of North America. In particular, males belonging to peoples such as the Buryats, Evens, Evenks, Kazakhs, Mongolians and Udegeys have high levels of M217.
One particular haplotype within Haplogroup C2 (M217) has received a great deal of attention, because of the possibility that it may represent direct patrilineal descent from Genghis Khan, one of his male-line ancestors, and/or other males belonging to the Borjigin clan. However, it is not certain that the Y-DNA haplogroup possessed by Genghis Khan was C2 (M217). Haplogroup C3c has been identified as a possible marker of the Manchu Aisin Gioro and is found in ten different ethnic minorities in northern China, and also found in Han Chinese.
Origin
Haplogroup C-M217 is believed to have originated approximately 7,100 to 16,700 years before present in eastern or central Asia. Its closest phylogenetic relatives are found in the general vicinity of South Asia, East Asia, or Oceania.
The extremely broad distribution of Haplogroup C-M217 Y-chromosomes, coupled with the fact that the ancestral paragroup C is not found among any of the modern Siberian or North American populations among whom Haplogroup C-M217 predominates, makes the determination of the geographical origin of the defining M217 mutation exceedingly difficult. The presence of Haplogroup C-M217 at a low frequency but relatively high diversity throughout East Asia and parts of Southeast Asia makes that region one likely source. In addition, the C-M217 haplotypes found with high frequency among North Asian populations appear to belong to a different genealogical branch from the C-M217 haplotypes found with low frequency among East and Southeast Asians, which suggests that the marginal presence of C-M217 among modern East and Southeast Asian populations may not be due to recent admixture from Northeast or Central Asia.
More precisely, haplogroup C-M217 is now divided into two primary subclades, C-F1067 and C-L1373. C-L1373 has been found often in populations from Central Asia through North Asia to the Americas, and rarely in individuals from some neighboring regions, such as Europe or East Asia. C-L1373 includes C-P39, which has been found with high frequency in samples of some indigenous North American populations, and C-M48, which is especially frequent among modern Tungusic peoples. The predominantly East Asian C-F1067 subsumes a major clade, C-F2613, and a minor clade, C-CTS4660. The minor clade C-CTS4660 has been found in China. The major clade C-F2613 has known representatives from China (including the Dai minority), Korea, Japan, Vietnam, Bangladesh, and Pakistan, and includes the populous subclades C-F845, C-CTS2657, and C-Z8440. C-M407, a notable subclade of C-CTS2657, has expanded in a post-Neolithic time frame to include large percentages of modern Buryat, Soyot, and Hamnigan males in Buryatia in addition to many Kalmyks and other Mongols (but only 2 or 0.67% of a sample of 300 Korean males).
Distribution
Haplogroup C-M217 is the modal haplogroup among Mongolians and most indigenous populations of the Russian Far East, such as the Northern Tungusic peoples, Koryaks, Itelmens, and Nivkhs. The subclade C-P39 is common among males of the indigenous North American peoples whose languages belong to the Na-Dené phylum. The frequency of Haplogroup C-M217 tends to be negatively correlated with distance from Mongolia and the Russian Far East, but it still comprises more than ten percent of the total Y-chromosome diversity among the Manchus, Koreans, Ainu, and some Turkic peoples of Central Asia although in a genetic study in 2004, haplogroup C-M217 was more frequent among Koreans than previously thought (16.5%). It is found at lower frequencies among Turkic peoples of Central Asia except Kazakhs. On the other hand, Mongols and Tungusic peoples have the highest frequencies. It is possible that this haplogroup might have spread into Turks during the Mongol invasions of 13th century as well as previous tribal leagues in the history. Beyond this range of high-to-moderate frequency, which contains mainly the northeast quadrant of Eurasia and the northwest quadrant of North America, Haplogroup C-M217 continues to be found at low frequencies, and it has even been found as far afield as Northwest Europe, Turkey, Pakistan, Nepal and adjacent regions of India, Vietnam, Maritime Southeast Asia, and the Wayuu people of South America.
In Japan, haplogroup C-M217 has been found relatively frequently among Ainus (2/16=12.5% or 1/4=25%) and among Japanese of the Kyūshū region (4/53=7.5% or 8/104=7.7%). In other regional Japanese populations, the frequency of haplogroup C-M217 hovers around 2%.
The frequency of Haplogroup C-M217 in samples of Han Chinese from various areas has ranged from 0% (0/27 Han from Guangxi) to 23.5% (8/34 Han from Xi'an), with the frequency of this haplogroup in several studies' pools of all Han Chinese samples ranging between 6.0% and 12.0%. C-M217 also has been found in samples of minority populations from central and southern China, such as Tujia from Jishou, Hunan (9/49 = 18.4%), Hani (6/34 = 17.6%), Miao (2/58 = 3.4%), She (1/34 = 2.9%), Yao (1/35 = 2.9% from Liannan, Guangdong), Tibetans (4/156 = 2.6%), and Taiwanese aborigines (1/48 = 2.1%). (Karafet 2010)(Xue 2006)(Gayden 2007)
In Vietnam, Haplogroup C-M217 has been found in 12.5% (6/48) of a sample of Vietnamese from Hanoi, Vietnam, 11.8% (9/76) of another sample of Kinh ("ethnic Vietnamese") from Hanoi, Vietnam, 8.5% (5/59) of a sample of Cham people from Binh Thuan, Vietnam, 8.3% (2/24) of another sample of Vietnamese from Hanoi, and in 4.3% (3/70) of a sample of Vietnamese from an unspecified location in Vietnam.(Karafet 2010)(He 2012) Haplogroup C-M217 has been found less frequently in other parts of Southeast Asia and nearby areas, including Laos (1/25 = 4.0% Lao from Luang Prabang), Java (1/37 = 2.7%), Nepal (2/77 = 2.6% general population of Kathmandu), Thailand (1/40 = 2.5% Thai, mostly sampled in Chiang Mai), the Philippines (1/48 = 2.1%, 1/64 = 1.6%), and Bali (1/641 = 0.2%). (Gayden 2007)(Karafet 2010)(He 2012)
Although C-M217 is generally found with only low frequency (<5%) in Tibet and Nepal, there may be an island of relatively high frequency of this haplogroup in Meghalaya, India. The indigenous tribes of this state of Northeast India, where they comprise the majority of the local population, speak Khasian languages or Tibeto-Burman languages. A study published in 2007 found C-M217(xM93, P39, M86) Y-DNA in 8.5% (6/71) of a sample of Garos, who primarily inhabit the Garo Hills in the western half of Meghalaya, and in 7.6% (27/353) of a pool of samples of eight Khasian tribes from the eastern half of Meghalaya (6/18 = 33.3% Nongtrai from the West Khasi Hills, 10/60 = 16.7% Lyngngam from the West Khasi Hills, 2/29 = 6.9% War-Khasi from the East Khasi Hills, 3/44 = 6.8% Pnar from the Jaintia Hills, 1/19 = 5.3% War-Jaintia from the Jaintia Hills, 3/87 = 3.4% Khynriam from the East Khasi Hills, 2/64 = 3.1% Maram from the West Khasi Hills, and 0/32 Bhoi from Ri-Bhoi District).(Reddy 2007)
Subclade distribution
The subclades of Haplogroup C-M217 with their defining mutation(s), according to the 2008 ISOGG tree:
C2-M217 Typical of Mongolians, Kazakhs, Buryats, Daurs, Kalmyks, Hazaras, Manchus, Sibes, Oroqens, Koryaks, and Itelmens; with a moderate distribution among other Tungusic peoples, Koreans, Ainus, Han Chinese, Vietnamese, Nivkhs, Altaians, Tuvinians, Uzbeks, Nogais, and Crimean Tatars
C2-M93 Observed sporadically in Japanese
C2-P39 Found in several indigenous peoples of North America, including some Na-Dené-, Algonquian-, or Siouan-speaking populations
C2-M48 Found frequently in Koryaks and sporadically among Evenks, Evens, and Yukaghirs
C2-M77 Typical of Northern Tungusic peoples, Kazakhs, Oirats, Kalmyks, Outer Mongolians, Yukaghirs, Nivkhs, Itelmens, and Udegeys, with a moderate distribution among other Southern Tungusic peoples, Inner Mongolians, Buryats, Tuvinians, Yakuts, Chukchi, Kyrgyz, Uyghurs, Uzbeks, Karakalpaks, and Tajiks
C2-M407 Found with high frequency in some samples of Buryats, Khamnigans, and Soyots, moderate frequency in Mongols and Kalmyks, and low frequency in Bai, Cambodian, Evenk, Han Chinese, Japanese, Korean, Manchu, Teleut, Tujia, Tuvinian, Uyghur, and Yakut populations
C2-P53.1 Found in about 10% of Xinjiang Sibe and with low frequency in Inner Mongolian Mongol and Evenk, Ningxia Hui, Xizang Tibetan, Xinjiang Uyghur, and Gansu Han
C2-P62
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
Research publications
The following research teams per their publications were represented in the creation of the YCC tree.
